197P Computational pathology pipeline enables quantification of intratumor heterogeneity and tumor-infiltrating lymphocyte score Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1016/j.iotech.2023.100656
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1016/j.iotech.2023.100656
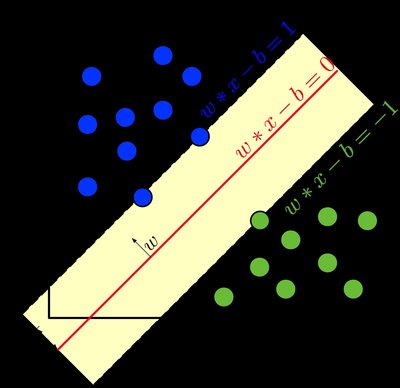
Tumor mutation burden, intratumor heterogeneity and the cancer related immune infiltrate are associated with response to target and immune therapies in several cancer types. The aim of this study is to establish a computational pathology (CPath) pipeline for investigating useful histopathological features in hematoxylin and eosin (H&E) whole slide images (WSIs). The pipeline allows for the segmentation and classification of nuclei. As an application, a machine learning approach is used to quantify intratumor heterogeneity (ITH) and tumor-infiltrating lymphocyte (TIL) scores. We randomly selected 178 invasive ductal carcinomas WSIs from the TCGA database to process. Image annotation and nuclei detection were performed on QuPath by a pathologist using StarDist and used to train a SVM-based nuclei type classifier. The classifier was trained to distinguish between tumor, lymphocyte, and stroma. A total of 113.211 nuclei images were extracted using OpenSlide and used to train an autoencoder for dimensionality reduction. The variability of the WSIs was computed by applying a statistical measure of dispersion to the feature vectors. The TIL score was computed as the average minimum distance between a tumor cell and its nearest lymphocyte neighbor. We used the mutation burden, the ITH, PAM50 classification, nuclear and histological grades data from [https://doi.org/10.1038/nm.3984] to validate our ITH score. The percentage of infiltrating lymphocytes inferred by the quanTIseq pipeline based on RNAseq data was used to validate the SVM model. The nuclei classifier achieved 88% accuracy on the test set and the 96%/92% sensitivity to tumor/lymphocyte on a validation dataset. The mean squared error of the autoencoder was 0.0004 in the test set. The ITH score was associated with the clonal number of a tumor (x2 = 8.8, p= 0.03). Finally, we observed a positive correlation between the TIL/tumor ratio inferred by the CPath pipeline and the total number of T lymphocytes inferred by quanTIseq (Spearman= 0. 34, p < 0.0001). We explored a CPath pipeline for detecting and classifying nuclei. The pipeline was used to compute ITH and TIL scores which correlated with validation data. Thus, validating our pipeline.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1016/j.iotech.2023.100656
- http://www.esmoiotech.org/article/S2590018823003428/pdf
- OA Status
- gold
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4389480801
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4389480801Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1016/j.iotech.2023.100656Digital Object Identifier
- Title
-
197P Computational pathology pipeline enables quantification of intratumor heterogeneity and tumor-infiltrating lymphocyte scoreWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-12-01Full publication date if available
- Authors
-
Daniel Guimarães Tiezzi, Antônio Augusto Fröhlich, Fernando Chahud, S. PagnotaList of authors in order
- Landing page
-
https://doi.org/10.1016/j.iotech.2023.100656Publisher landing page
- PDF URL
-
https://www.esmoiotech.org/article/S2590018823003428/pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.esmoiotech.org/article/S2590018823003428/pdfDirect OA link when available
- Concepts
-
Digital pathology, Artificial intelligence, Support vector machine, Tumor-infiltrating lymphocytes, Lymphocyte, Computer science, Classifier (UML), Pathology, Pattern recognition (psychology), Medicine, Immune system, Immunology, CD8Top concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4389480801 |
|---|---|
| doi | https://doi.org/10.1016/j.iotech.2023.100656 |
| ids.doi | https://doi.org/10.1016/j.iotech.2023.100656 |
| ids.openalex | https://openalex.org/W4389480801 |
| fwci | 0.0 |
| type | article |
| title | 197P Computational pathology pipeline enables quantification of intratumor heterogeneity and tumor-infiltrating lymphocyte score |
| biblio.issue | |
| biblio.volume | 20 |
| biblio.last_page | 100656 |
| biblio.first_page | 100656 |
| topics[0].id | https://openalex.org/T12422 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9976999759674072 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2741 |
| topics[0].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[0].display_name | Radiomics and Machine Learning in Medical Imaging |
| topics[1].id | https://openalex.org/T10862 |
| topics[1].field.id | https://openalex.org/fields/17 |
| topics[1].field.display_name | Computer Science |
| topics[1].score | 0.9857000112533569 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1702 |
| topics[1].subfield.display_name | Artificial Intelligence |
| topics[1].display_name | AI in cancer detection |
| topics[2].id | https://openalex.org/T13110 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9746999740600586 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2746 |
| topics[2].subfield.display_name | Surgery |
| topics[2].display_name | Colorectal and Anal Carcinomas |
| is_xpac | False |
| apc_list.value | 2750 |
| apc_list.currency | EUR |
| apc_list.value_usd | 2965 |
| apc_paid.value | 2750 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 2965 |
| concepts[0].id | https://openalex.org/C2777522853 |
| concepts[0].level | 2 |
| concepts[0].score | 0.626690149307251 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q5276128 |
| concepts[0].display_name | Digital pathology |
| concepts[1].id | https://openalex.org/C154945302 |
| concepts[1].level | 1 |
| concepts[1].score | 0.5494798421859741 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[1].display_name | Artificial intelligence |
| concepts[2].id | https://openalex.org/C12267149 |
| concepts[2].level | 2 |
| concepts[2].score | 0.44839537143707275 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q282453 |
| concepts[2].display_name | Support vector machine |
| concepts[3].id | https://openalex.org/C2778326572 |
| concepts[3].level | 4 |
| concepts[3].score | 0.44121047854423523 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7852668 |
| concepts[3].display_name | Tumor-infiltrating lymphocytes |
| concepts[4].id | https://openalex.org/C2777761686 |
| concepts[4].level | 2 |
| concepts[4].score | 0.43992099165916443 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q715347 |
| concepts[4].display_name | Lymphocyte |
| concepts[5].id | https://openalex.org/C41008148 |
| concepts[5].level | 0 |
| concepts[5].score | 0.41343623399734497 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[5].display_name | Computer science |
| concepts[6].id | https://openalex.org/C95623464 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4103371798992157 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1096149 |
| concepts[6].display_name | Classifier (UML) |
| concepts[7].id | https://openalex.org/C142724271 |
| concepts[7].level | 1 |
| concepts[7].score | 0.40408483147621155 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[7].display_name | Pathology |
| concepts[8].id | https://openalex.org/C153180895 |
| concepts[8].level | 2 |
| concepts[8].score | 0.34276682138442993 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7148389 |
| concepts[8].display_name | Pattern recognition (psychology) |
| concepts[9].id | https://openalex.org/C71924100 |
| concepts[9].level | 0 |
| concepts[9].score | 0.3137190639972687 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[9].display_name | Medicine |
| concepts[10].id | https://openalex.org/C8891405 |
| concepts[10].level | 2 |
| concepts[10].score | 0.2521374523639679 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q1059 |
| concepts[10].display_name | Immune system |
| concepts[11].id | https://openalex.org/C203014093 |
| concepts[11].level | 1 |
| concepts[11].score | 0.11667248606681824 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[11].display_name | Immunology |
| concepts[12].id | https://openalex.org/C167672396 |
| concepts[12].level | 3 |
| concepts[12].score | 0.10649305582046509 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q417800 |
| concepts[12].display_name | CD8 |
| keywords[0].id | https://openalex.org/keywords/digital-pathology |
| keywords[0].score | 0.626690149307251 |
| keywords[0].display_name | Digital pathology |
| keywords[1].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[1].score | 0.5494798421859741 |
| keywords[1].display_name | Artificial intelligence |
| keywords[2].id | https://openalex.org/keywords/support-vector-machine |
| keywords[2].score | 0.44839537143707275 |
| keywords[2].display_name | Support vector machine |
| keywords[3].id | https://openalex.org/keywords/tumor-infiltrating-lymphocytes |
| keywords[3].score | 0.44121047854423523 |
| keywords[3].display_name | Tumor-infiltrating lymphocytes |
| keywords[4].id | https://openalex.org/keywords/lymphocyte |
| keywords[4].score | 0.43992099165916443 |
| keywords[4].display_name | Lymphocyte |
| keywords[5].id | https://openalex.org/keywords/computer-science |
| keywords[5].score | 0.41343623399734497 |
| keywords[5].display_name | Computer science |
| keywords[6].id | https://openalex.org/keywords/classifier |
| keywords[6].score | 0.4103371798992157 |
| keywords[6].display_name | Classifier (UML) |
| keywords[7].id | https://openalex.org/keywords/pathology |
| keywords[7].score | 0.40408483147621155 |
| keywords[7].display_name | Pathology |
| keywords[8].id | https://openalex.org/keywords/pattern-recognition |
| keywords[8].score | 0.34276682138442993 |
| keywords[8].display_name | Pattern recognition (psychology) |
| keywords[9].id | https://openalex.org/keywords/medicine |
| keywords[9].score | 0.3137190639972687 |
| keywords[9].display_name | Medicine |
| keywords[10].id | https://openalex.org/keywords/immune-system |
| keywords[10].score | 0.2521374523639679 |
| keywords[10].display_name | Immune system |
| keywords[11].id | https://openalex.org/keywords/immunology |
| keywords[11].score | 0.11667248606681824 |
| keywords[11].display_name | Immunology |
| keywords[12].id | https://openalex.org/keywords/cd8 |
| keywords[12].score | 0.10649305582046509 |
| keywords[12].display_name | CD8 |
| language | en |
| locations[0].id | doi:10.1016/j.iotech.2023.100656 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210240197 |
| locations[0].source.issn | 2590-0188 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2590-0188 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Immuno-Oncology Technology |
| locations[0].source.host_organization | https://openalex.org/P4310320990 |
| locations[0].source.host_organization_name | Elsevier BV |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320990 |
| locations[0].source.host_organization_lineage_names | Elsevier BV |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | http://www.esmoiotech.org/article/S2590018823003428/pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Immuno-Oncology and Technology |
| locations[0].landing_page_url | https://doi.org/10.1016/j.iotech.2023.100656 |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5008447927 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-2660-0093 |
| authorships[0].author.display_name | Daniel Guimarães Tiezzi |
| authorships[0].countries | BR |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210156537 |
| authorships[0].affiliations[0].raw_affiliation_string | Breast Disease Division, HC-FMRP-USP - Hospital das Clínicas da Faculdade de Medicina de Ribeirao, Ribeirão Preto, Brazil |
| authorships[0].institutions[0].id | https://openalex.org/I4210156537 |
| authorships[0].institutions[0].ror | https://ror.org/04n6fhj26 |
| authorships[0].institutions[0].type | healthcare |
| authorships[0].institutions[0].lineage | https://openalex.org/I17974374, https://openalex.org/I4210156537 |
| authorships[0].institutions[0].country_code | BR |
| authorships[0].institutions[0].display_name | Clinics Hospital of Ribeirão Preto |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | D.G. Tiezzi |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Breast Disease Division, HC-FMRP-USP - Hospital das Clínicas da Faculdade de Medicina de Ribeirao, Ribeirão Preto, Brazil |
| authorships[1].author.id | https://openalex.org/A5039916641 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4063-1339 |
| authorships[1].author.display_name | Antônio Augusto Fröhlich |
| authorships[1].countries | BR |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4104125 |
| authorships[1].affiliations[0].raw_affiliation_string | Mathematics, Federal University of Santa Catarina UFSC, Araranguá, Brazil |
| authorships[1].institutions[0].id | https://openalex.org/I4104125 |
| authorships[1].institutions[0].ror | https://ror.org/041akq887 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I4104125 |
| authorships[1].institutions[0].country_code | BR |
| authorships[1].institutions[0].display_name | Universidade Federal de Santa Catarina |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | A. Fröhlich |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Mathematics, Federal University of Santa Catarina UFSC, Araranguá, Brazil |
| authorships[2].author.id | https://openalex.org/A5082227062 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-8099-9648 |
| authorships[2].author.display_name | Fernando Chahud |
| authorships[2].countries | BR |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210156537 |
| authorships[2].affiliations[0].raw_affiliation_string | Pathology, Faculdade de Medicina de Ribeirão Preto, Ribeirão Preto, Brazil |
| authorships[2].institutions[0].id | https://openalex.org/I4210156537 |
| authorships[2].institutions[0].ror | https://ror.org/04n6fhj26 |
| authorships[2].institutions[0].type | healthcare |
| authorships[2].institutions[0].lineage | https://openalex.org/I17974374, https://openalex.org/I4210156537 |
| authorships[2].institutions[0].country_code | BR |
| authorships[2].institutions[0].display_name | Clinics Hospital of Ribeirão Preto |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | F. Chahud |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Pathology, Faculdade de Medicina de Ribeirão Preto, Ribeirão Preto, Brazil |
| authorships[3].author.id | https://openalex.org/A5093447997 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | S. Pagnota |
| authorships[3].countries | IT |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I16337185 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Science and Technology (DST), University of Sannio RCOST, Benevento, Italy |
| authorships[3].institutions[0].id | https://openalex.org/I16337185 |
| authorships[3].institutions[0].ror | https://ror.org/04vc81p87 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I16337185 |
| authorships[3].institutions[0].country_code | IT |
| authorships[3].institutions[0].display_name | University of Sannio |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | S. Pagnota |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Science and Technology (DST), University of Sannio RCOST, Benevento, Italy |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | http://www.esmoiotech.org/article/S2590018823003428/pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | 197P Computational pathology pipeline enables quantification of intratumor heterogeneity and tumor-infiltrating lymphocyte score |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12422 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9976999759674072 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2741 |
| primary_topic.subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| primary_topic.display_name | Radiomics and Machine Learning in Medical Imaging |
| related_works | https://openalex.org/W2090763504, https://openalex.org/W1975767702, https://openalex.org/W148178222, https://openalex.org/W2104657898, https://openalex.org/W1948992892, https://openalex.org/W3128797991, https://openalex.org/W1886884218, https://openalex.org/W1910826599, https://openalex.org/W1980100242, https://openalex.org/W2530420969 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1016/j.iotech.2023.100656 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210240197 |
| best_oa_location.source.issn | 2590-0188 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2590-0188 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Immuno-Oncology Technology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320990 |
| best_oa_location.source.host_organization_name | Elsevier BV |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320990 |
| best_oa_location.source.host_organization_lineage_names | Elsevier BV |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | http://www.esmoiotech.org/article/S2590018823003428/pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Immuno-Oncology and Technology |
| best_oa_location.landing_page_url | https://doi.org/10.1016/j.iotech.2023.100656 |
| primary_location.id | doi:10.1016/j.iotech.2023.100656 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210240197 |
| primary_location.source.issn | 2590-0188 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2590-0188 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Immuno-Oncology Technology |
| primary_location.source.host_organization | https://openalex.org/P4310320990 |
| primary_location.source.host_organization_name | Elsevier BV |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320990 |
| primary_location.source.host_organization_lineage_names | Elsevier BV |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | http://www.esmoiotech.org/article/S2590018823003428/pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Immuno-Oncology and Technology |
| primary_location.landing_page_url | https://doi.org/10.1016/j.iotech.2023.100656 |
| publication_date | 2023-12-01 |
| publication_year | 2023 |
| referenced_works_count | 0 |
| abstract_inverted_index.< | 305 |
| abstract_inverted_index.= | 272 |
| abstract_inverted_index.A | 128 |
| abstract_inverted_index.T | 296 |
| abstract_inverted_index.a | 32, 64, 104, 112, 156, 176, 243, 269, 279, 309 |
| abstract_inverted_index.p | 304 |
| abstract_inverted_index.0. | 302 |
| abstract_inverted_index.As | 61 |
| abstract_inverted_index.We | 80, 184, 307 |
| abstract_inverted_index.an | 62, 142 |
| abstract_inverted_index.as | 170 |
| abstract_inverted_index.by | 103, 154, 211, 287, 299 |
| abstract_inverted_index.in | 20, 42, 255 |
| abstract_inverted_index.is | 29, 68 |
| abstract_inverted_index.of | 26, 59, 130, 149, 159, 207, 250, 268, 295 |
| abstract_inverted_index.on | 101, 216, 232, 242 |
| abstract_inverted_index.p= | 274 |
| abstract_inverted_index.to | 15, 30, 70, 92, 110, 121, 140, 161, 200, 221, 240, 321 |
| abstract_inverted_index.we | 277 |
| abstract_inverted_index.(x2 | 271 |
| abstract_inverted_index.178 | 83 |
| abstract_inverted_index.34, | 303 |
| abstract_inverted_index.88% | 230 |
| abstract_inverted_index.ITH | 203, 260, 323 |
| abstract_inverted_index.SVM | 224 |
| abstract_inverted_index.TIL | 166, 325 |
| abstract_inverted_index.The | 24, 51, 117, 147, 165, 205, 226, 246, 259, 317 |
| abstract_inverted_index.aim | 25 |
| abstract_inverted_index.and | 5, 17, 44, 57, 75, 96, 108, 126, 138, 179, 194, 236, 291, 314, 324 |
| abstract_inverted_index.are | 11 |
| abstract_inverted_index.for | 37, 54, 144, 312 |
| abstract_inverted_index.its | 180 |
| abstract_inverted_index.our | 202, 334 |
| abstract_inverted_index.set | 235 |
| abstract_inverted_index.the | 6, 55, 89, 150, 162, 171, 186, 189, 212, 223, 233, 237, 251, 256, 265, 283, 288, 292 |
| abstract_inverted_index.was | 119, 152, 168, 219, 253, 262, 319 |
| abstract_inverted_index.8.8, | 273 |
| abstract_inverted_index.ITH, | 190 |
| abstract_inverted_index.TCGA | 90 |
| abstract_inverted_index.WSIs | 87, 151 |
| abstract_inverted_index.cell | 178 |
| abstract_inverted_index.data | 197, 218 |
| abstract_inverted_index.from | 88, 198 |
| abstract_inverted_index.mean | 247 |
| abstract_inverted_index.set. | 258 |
| abstract_inverted_index.test | 234, 257 |
| abstract_inverted_index.this | 27 |
| abstract_inverted_index.type | 115 |
| abstract_inverted_index.used | 69, 109, 139, 185, 220, 320 |
| abstract_inverted_index.were | 99, 134 |
| abstract_inverted_index.with | 13, 264, 329 |
| abstract_inverted_index.(H&E) | 46 |
| abstract_inverted_index.(ITH) | 74 |
| abstract_inverted_index.(TIL) | 78 |
| abstract_inverted_index.CPath | 289, 310 |
| abstract_inverted_index.Image | 94 |
| abstract_inverted_index.PAM50 | 191 |
| abstract_inverted_index.Thus, | 332 |
| abstract_inverted_index.Tumor | 0 |
| abstract_inverted_index.based | 215 |
| abstract_inverted_index.data. | 331 |
| abstract_inverted_index.eosin | 45 |
| abstract_inverted_index.error | 249 |
| abstract_inverted_index.ratio | 285 |
| abstract_inverted_index.score | 167, 261 |
| abstract_inverted_index.slide | 48 |
| abstract_inverted_index.study | 28 |
| abstract_inverted_index.total | 129, 293 |
| abstract_inverted_index.train | 111, 141 |
| abstract_inverted_index.tumor | 177, 270 |
| abstract_inverted_index.using | 106, 136 |
| abstract_inverted_index.which | 327 |
| abstract_inverted_index.whole | 47 |
| abstract_inverted_index.0.0004 | 254 |
| abstract_inverted_index.0.03). | 275 |
| abstract_inverted_index.QuPath | 102 |
| abstract_inverted_index.RNAseq | 217 |
| abstract_inverted_index.allows | 53 |
| abstract_inverted_index.cancer | 7, 22 |
| abstract_inverted_index.clonal | 266 |
| abstract_inverted_index.ductal | 85 |
| abstract_inverted_index.grades | 196 |
| abstract_inverted_index.images | 49, 133 |
| abstract_inverted_index.immune | 9, 18 |
| abstract_inverted_index.model. | 225 |
| abstract_inverted_index.nuclei | 97, 114, 132, 227 |
| abstract_inverted_index.number | 267, 294 |
| abstract_inverted_index.score. | 204 |
| abstract_inverted_index.scores | 326 |
| abstract_inverted_index.target | 16 |
| abstract_inverted_index.tumor, | 124 |
| abstract_inverted_index.types. | 23 |
| abstract_inverted_index.useful | 39 |
| abstract_inverted_index.(CPath) | 35 |
| abstract_inverted_index.(WSIs). | 50 |
| abstract_inverted_index.113.211 | 131 |
| abstract_inverted_index.96%/92% | 238 |
| abstract_inverted_index.average | 172 |
| abstract_inverted_index.between | 123, 175, 282 |
| abstract_inverted_index.burden, | 2, 188 |
| abstract_inverted_index.compute | 322 |
| abstract_inverted_index.feature | 163 |
| abstract_inverted_index.machine | 65 |
| abstract_inverted_index.measure | 158 |
| abstract_inverted_index.minimum | 173 |
| abstract_inverted_index.nearest | 181 |
| abstract_inverted_index.nuclear | 193 |
| abstract_inverted_index.nuclei. | 60, 316 |
| abstract_inverted_index.related | 8 |
| abstract_inverted_index.scores. | 79 |
| abstract_inverted_index.several | 21 |
| abstract_inverted_index.squared | 248 |
| abstract_inverted_index.stroma. | 127 |
| abstract_inverted_index.trained | 120 |
| abstract_inverted_index.0.0001). | 306 |
| abstract_inverted_index.Finally, | 276 |
| abstract_inverted_index.StarDist | 107 |
| abstract_inverted_index.accuracy | 231 |
| abstract_inverted_index.achieved | 229 |
| abstract_inverted_index.applying | 155 |
| abstract_inverted_index.approach | 67 |
| abstract_inverted_index.computed | 153, 169 |
| abstract_inverted_index.database | 91 |
| abstract_inverted_index.dataset. | 245 |
| abstract_inverted_index.distance | 174 |
| abstract_inverted_index.explored | 308 |
| abstract_inverted_index.features | 41 |
| abstract_inverted_index.inferred | 210, 286, 298 |
| abstract_inverted_index.invasive | 84 |
| abstract_inverted_index.learning | 66 |
| abstract_inverted_index.mutation | 1, 187 |
| abstract_inverted_index.observed | 278 |
| abstract_inverted_index.pipeline | 36, 52, 214, 290, 311, 318 |
| abstract_inverted_index.positive | 280 |
| abstract_inverted_index.process. | 93 |
| abstract_inverted_index.quantify | 71 |
| abstract_inverted_index.randomly | 81 |
| abstract_inverted_index.response | 14 |
| abstract_inverted_index.selected | 82 |
| abstract_inverted_index.validate | 201, 222 |
| abstract_inverted_index.vectors. | 164 |
| abstract_inverted_index.OpenSlide | 137 |
| abstract_inverted_index.SVM-based | 113 |
| abstract_inverted_index.TIL/tumor | 284 |
| abstract_inverted_index.detecting | 313 |
| abstract_inverted_index.detection | 98 |
| abstract_inverted_index.establish | 31 |
| abstract_inverted_index.extracted | 135 |
| abstract_inverted_index.neighbor. | 183 |
| abstract_inverted_index.pathology | 34 |
| abstract_inverted_index.performed | 100 |
| abstract_inverted_index.pipeline. | 335 |
| abstract_inverted_index.quanTIseq | 213, 300 |
| abstract_inverted_index.therapies | 19 |
| abstract_inverted_index.(Spearman= | 301 |
| abstract_inverted_index.annotation | 95 |
| abstract_inverted_index.associated | 12, 263 |
| abstract_inverted_index.carcinomas | 86 |
| abstract_inverted_index.classifier | 118, 228 |
| abstract_inverted_index.correlated | 328 |
| abstract_inverted_index.dispersion | 160 |
| abstract_inverted_index.infiltrate | 10 |
| abstract_inverted_index.intratumor | 3, 72 |
| abstract_inverted_index.lymphocyte | 77, 182 |
| abstract_inverted_index.percentage | 206 |
| abstract_inverted_index.reduction. | 146 |
| abstract_inverted_index.validating | 333 |
| abstract_inverted_index.validation | 244, 330 |
| abstract_inverted_index.autoencoder | 143, 252 |
| abstract_inverted_index.classifier. | 116 |
| abstract_inverted_index.classifying | 315 |
| abstract_inverted_index.correlation | 281 |
| abstract_inverted_index.distinguish | 122 |
| abstract_inverted_index.hematoxylin | 43 |
| abstract_inverted_index.lymphocyte, | 125 |
| abstract_inverted_index.lymphocytes | 209, 297 |
| abstract_inverted_index.pathologist | 105 |
| abstract_inverted_index.sensitivity | 239 |
| abstract_inverted_index.statistical | 157 |
| abstract_inverted_index.variability | 148 |
| abstract_inverted_index.application, | 63 |
| abstract_inverted_index.histological | 195 |
| abstract_inverted_index.infiltrating | 208 |
| abstract_inverted_index.segmentation | 56 |
| abstract_inverted_index.computational | 33 |
| abstract_inverted_index.heterogeneity | 4, 73 |
| abstract_inverted_index.investigating | 38 |
| abstract_inverted_index.classification | 58 |
| abstract_inverted_index.dimensionality | 145 |
| abstract_inverted_index.classification, | 192 |
| abstract_inverted_index.tumor/lymphocyte | 241 |
| abstract_inverted_index.histopathological | 40 |
| abstract_inverted_index.tumor-infiltrating | 76 |
| abstract_inverted_index.[https://doi.org/10.1038/nm.3984] | 199 |
| cited_by_percentile_year | |
| countries_distinct_count | 2 |
| institutions_distinct_count | 4 |
| citation_normalized_percentile.value | 0.38715268 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |