A Gene Based Optimally Weighted Combination Test Using GWAS Summary Data Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.21203/rs.3.rs-1720857/v1
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.21203/rs.3.rs-1720857/v1
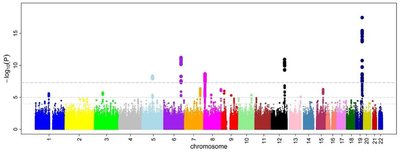
Background: Gene-based association tests provide a useful alternative and complement to the usual single marker association tests, especially in genome-wide association studies (GWAS). The way of weighting for variants in a gene plays an important role in boosting the power of a gene-based association test. Appropriate weights can boost statistical power, especially when detecting genetic variants with weak effects on a trait. However, the effect sizes or directions of causal genetic variants in real data are usually unknown, driving a need for a flexible yet robust methodology. Furthermore, access to individual-level data is often limited, while thousands of GWAS summary data are publicly and freely available. Results: To resolve these limitations, we propose an optimally weighted combination (OWC) test based on summary statistics from GWAS data. Several traditional methods including burden test, weighted sum of squared score test [SSU], weighted sum statistic [WSS], SNP-set Kernel Association Test [SKAT], and the score test are special cases of OWC. To evaluate the performance of OWC, we perform extensive simulation studies. Results of simulation studies demonstrate that OWC outperforms several existing popular methods. We further show that OWC outperforms comparison methods in real-world data analyses using schizophrenia GWAS summary data and a fasting glucose GWAS meta-analysis data. The proposed method is implemented in an R package available at https://github.com/Jianjun-CN/R-package-for-OWC. Conclusions: We propose a novel gene-based association test that incorporates four different weighting schemes (two constant weights and two weights proportional to normal statistic Z) and includes several popular methods as its special cases. Results of the simulation studies and real data analyses illustrate that the proposed test, OWC, outperforms comparable methods in most scenarios. These demonstrate that OWC is a useful tool that adapts to the underlying biological model for a disease by optimally weighting variants.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.21203/rs.3.rs-1720857/v1
- https://www.researchsquare.com/article/rs-1720857/latest.pdf
- OA Status
- green
- References
- 30
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4283708494
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4283708494Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.21203/rs.3.rs-1720857/v1Digital Object Identifier
- Title
-
A Gene Based Optimally Weighted Combination Test Using GWAS Summary DataWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-06-29Full publication date if available
- Authors
-
Jianjun Zhang, Xiaoyu Liang, Samantha Gonzales, Jianguo Liu, Xiaoyi Gao, Xuexia WangList of authors in order
- Landing page
-
https://doi.org/10.21203/rs.3.rs-1720857/v1Publisher landing page
- PDF URL
-
https://www.researchsquare.com/article/rs-1720857/latest.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.researchsquare.com/article/rs-1720857/latest.pdfDirect OA link when available
- Concepts
-
Genome-wide association study, Computer science, Weighting, Statistic, Genetic association, Score test, Data mining, Association test, Statistics, Statistical hypothesis testing, Mathematics, Single-nucleotide polymorphism, Medicine, Biology, Gene, Genetics, Radiology, GenotypeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
30Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4283708494 |
|---|---|
| doi | https://doi.org/10.21203/rs.3.rs-1720857/v1 |
| ids.doi | https://doi.org/10.21203/rs.3.rs-1720857/v1 |
| ids.openalex | https://openalex.org/W4283708494 |
| fwci | 0.0 |
| type | preprint |
| title | A Gene Based Optimally Weighted Combination Test Using GWAS Summary Data |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10261 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9993000030517578 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic Associations and Epidemiology |
| topics[1].id | https://openalex.org/T10845 |
| topics[1].field.id | https://openalex.org/fields/26 |
| topics[1].field.display_name | Mathematics |
| topics[1].score | 0.9643999934196472 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2613 |
| topics[1].subfield.display_name | Statistics and Probability |
| topics[1].display_name | Advanced Causal Inference Techniques |
| topics[2].id | https://openalex.org/T11577 |
| topics[2].field.id | https://openalex.org/fields/32 |
| topics[2].field.display_name | Psychology |
| topics[2].score | 0.9398000240325928 |
| topics[2].domain.id | https://openalex.org/domains/2 |
| topics[2].domain.display_name | Social Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/3205 |
| topics[2].subfield.display_name | Experimental and Cognitive Psychology |
| topics[2].display_name | Cognitive Abilities and Testing |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7049509882926941 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C41008148 |
| concepts[1].level | 0 |
| concepts[1].score | 0.5717056393623352 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[1].display_name | Computer science |
| concepts[2].id | https://openalex.org/C183115368 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5550925731658936 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q856577 |
| concepts[2].display_name | Weighting |
| concepts[3].id | https://openalex.org/C89128539 |
| concepts[3].level | 2 |
| concepts[3].score | 0.513533353805542 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1949963 |
| concepts[3].display_name | Statistic |
| concepts[4].id | https://openalex.org/C186413461 |
| concepts[4].level | 5 |
| concepts[4].score | 0.4690285921096802 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[4].display_name | Genetic association |
| concepts[5].id | https://openalex.org/C103463560 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4595434069633484 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q4456449 |
| concepts[5].display_name | Score test |
| concepts[6].id | https://openalex.org/C124101348 |
| concepts[6].level | 1 |
| concepts[6].score | 0.45797985792160034 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q172491 |
| concepts[6].display_name | Data mining |
| concepts[7].id | https://openalex.org/C2994538360 |
| concepts[7].level | 5 |
| concepts[7].score | 0.45350176095962524 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q774081 |
| concepts[7].display_name | Association test |
| concepts[8].id | https://openalex.org/C105795698 |
| concepts[8].level | 1 |
| concepts[8].score | 0.3705565333366394 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[8].display_name | Statistics |
| concepts[9].id | https://openalex.org/C87007009 |
| concepts[9].level | 2 |
| concepts[9].score | 0.3553656041622162 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q210832 |
| concepts[9].display_name | Statistical hypothesis testing |
| concepts[10].id | https://openalex.org/C33923547 |
| concepts[10].level | 0 |
| concepts[10].score | 0.24104753136634827 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[10].display_name | Mathematics |
| concepts[11].id | https://openalex.org/C153209595 |
| concepts[11].level | 4 |
| concepts[11].score | 0.2330317199230194 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[11].display_name | Single-nucleotide polymorphism |
| concepts[12].id | https://openalex.org/C71924100 |
| concepts[12].level | 0 |
| concepts[12].score | 0.136897474527359 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[12].display_name | Medicine |
| concepts[13].id | https://openalex.org/C86803240 |
| concepts[13].level | 0 |
| concepts[13].score | 0.11371412873268127 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[13].display_name | Biology |
| concepts[14].id | https://openalex.org/C104317684 |
| concepts[14].level | 2 |
| concepts[14].score | 0.09917765855789185 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[14].display_name | Gene |
| concepts[15].id | https://openalex.org/C54355233 |
| concepts[15].level | 1 |
| concepts[15].score | 0.08954352140426636 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[15].display_name | Genetics |
| concepts[16].id | https://openalex.org/C126838900 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q77604 |
| concepts[16].display_name | Radiology |
| concepts[17].id | https://openalex.org/C135763542 |
| concepts[17].level | 3 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[17].display_name | Genotype |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.7049509882926941 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/computer-science |
| keywords[1].score | 0.5717056393623352 |
| keywords[1].display_name | Computer science |
| keywords[2].id | https://openalex.org/keywords/weighting |
| keywords[2].score | 0.5550925731658936 |
| keywords[2].display_name | Weighting |
| keywords[3].id | https://openalex.org/keywords/statistic |
| keywords[3].score | 0.513533353805542 |
| keywords[3].display_name | Statistic |
| keywords[4].id | https://openalex.org/keywords/genetic-association |
| keywords[4].score | 0.4690285921096802 |
| keywords[4].display_name | Genetic association |
| keywords[5].id | https://openalex.org/keywords/score-test |
| keywords[5].score | 0.4595434069633484 |
| keywords[5].display_name | Score test |
| keywords[6].id | https://openalex.org/keywords/data-mining |
| keywords[6].score | 0.45797985792160034 |
| keywords[6].display_name | Data mining |
| keywords[7].id | https://openalex.org/keywords/association-test |
| keywords[7].score | 0.45350176095962524 |
| keywords[7].display_name | Association test |
| keywords[8].id | https://openalex.org/keywords/statistics |
| keywords[8].score | 0.3705565333366394 |
| keywords[8].display_name | Statistics |
| keywords[9].id | https://openalex.org/keywords/statistical-hypothesis-testing |
| keywords[9].score | 0.3553656041622162 |
| keywords[9].display_name | Statistical hypothesis testing |
| keywords[10].id | https://openalex.org/keywords/mathematics |
| keywords[10].score | 0.24104753136634827 |
| keywords[10].display_name | Mathematics |
| keywords[11].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[11].score | 0.2330317199230194 |
| keywords[11].display_name | Single-nucleotide polymorphism |
| keywords[12].id | https://openalex.org/keywords/medicine |
| keywords[12].score | 0.136897474527359 |
| keywords[12].display_name | Medicine |
| keywords[13].id | https://openalex.org/keywords/biology |
| keywords[13].score | 0.11371412873268127 |
| keywords[13].display_name | Biology |
| keywords[14].id | https://openalex.org/keywords/gene |
| keywords[14].score | 0.09917765855789185 |
| keywords[14].display_name | Gene |
| keywords[15].id | https://openalex.org/keywords/genetics |
| keywords[15].score | 0.08954352140426636 |
| keywords[15].display_name | Genetics |
| language | en |
| locations[0].id | doi:10.21203/rs.3.rs-1720857/v1 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402450 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Research Square (Research Square) |
| locations[0].source.host_organization | https://openalex.org/I4210096694 |
| locations[0].source.host_organization_name | Research Square (United States) |
| locations[0].source.host_organization_lineage | https://openalex.org/I4210096694 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.researchsquare.com/article/rs-1720857/latest.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.21203/rs.3.rs-1720857/v1 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5100358183 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-0045-8316 |
| authorships[0].author.display_name | Jianjun Zhang |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I123534392 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I123534392 |
| authorships[0].affiliations[1].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[0].institutions[0].id | https://openalex.org/I123534392 |
| authorships[0].institutions[0].ror | https://ror.org/00v97ad02 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I123534392 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of North Texas |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jianjun Zhang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA, Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[1].author.id | https://openalex.org/A5077833737 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-7796-2441 |
| authorships[1].author.display_name | Xiaoyu Liang |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I160606119 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Preventive Medicine, University of Tennessee Health Science Center, 66 N. Pauline Street, Memphis, TN 38105, USA. |
| authorships[1].institutions[0].id | https://openalex.org/I160606119 |
| authorships[1].institutions[0].ror | https://ror.org/0011qv509 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I160606119 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of Tennessee Health Science Center |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Xiaoyu Liang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Preventive Medicine, University of Tennessee Health Science Center, 66 N. Pauline Street, Memphis, TN 38105, USA. |
| authorships[2].author.id | https://openalex.org/A5059133513 |
| authorships[2].author.orcid | https://orcid.org/0009-0001-9212-8102 |
| authorships[2].author.display_name | Samantha Gonzales |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I123534392 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I123534392 |
| authorships[2].affiliations[1].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[2].institutions[0].id | https://openalex.org/I123534392 |
| authorships[2].institutions[0].ror | https://ror.org/00v97ad02 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I123534392 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of North Texas |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Samantha Gonzales |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA, Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[3].author.id | https://openalex.org/A5100333038 |
| authorships[3].author.orcid | https://orcid.org/0000-0001-7748-0960 |
| authorships[3].author.display_name | Jianguo Liu |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I123534392 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I123534392 |
| authorships[3].affiliations[1].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[3].institutions[0].id | https://openalex.org/I123534392 |
| authorships[3].institutions[0].ror | https://ror.org/00v97ad02 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I123534392 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of North Texas |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jianguo Liu |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA, Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[4].author.id | https://openalex.org/A5102721077 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-7251-8218 |
| authorships[4].author.display_name | Xiaoyi Gao |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I52357470 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Ophthalmology and Visual Science, Department of Biomedical informatics, Division of Human Genetics, Ohio State University, 915 Olentangy River Road, Columbus, OH 43212, USA. |
| authorships[4].institutions[0].id | https://openalex.org/I52357470 |
| authorships[4].institutions[0].ror | https://ror.org/00rs6vg23 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I52357470 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | The Ohio State University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Xiaoyi Raymond Gao |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Ophthalmology and Visual Science, Department of Biomedical informatics, Division of Human Genetics, Ohio State University, 915 Olentangy River Road, Columbus, OH 43212, USA. |
| authorships[5].author.id | https://openalex.org/A5101511802 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-0613-2608 |
| authorships[5].author.display_name | Xuexia Wang |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I123534392 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I123534392 |
| authorships[5].affiliations[1].raw_affiliation_string | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| authorships[5].institutions[0].id | https://openalex.org/I123534392 |
| authorships[5].institutions[0].ror | https://ror.org/00v97ad02 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I123534392 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | University of North Texas |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | Xuexia Wang |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA, Department of Mathematics, University of North Texas, 225 Avenue E, Denton, TX 76201, USA. |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.researchsquare.com/article/rs-1720857/latest.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | A Gene Based Optimally Weighted Combination Test Using GWAS Summary Data |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10261 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9993000030517578 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic Associations and Epidemiology |
| related_works | https://openalex.org/W1971424770, https://openalex.org/W2111842731, https://openalex.org/W2066467839, https://openalex.org/W2594299395, https://openalex.org/W2771541573, https://openalex.org/W2765794416, https://openalex.org/W1486217206, https://openalex.org/W2170506381, https://openalex.org/W2794350630, https://openalex.org/W2950093545 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.21203/rs.3.rs-1720857/v1 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402450 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Research Square (Research Square) |
| best_oa_location.source.host_organization | https://openalex.org/I4210096694 |
| best_oa_location.source.host_organization_name | Research Square (United States) |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I4210096694 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.researchsquare.com/article/rs-1720857/latest.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.21203/rs.3.rs-1720857/v1 |
| primary_location.id | doi:10.21203/rs.3.rs-1720857/v1 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402450 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Research Square (Research Square) |
| primary_location.source.host_organization | https://openalex.org/I4210096694 |
| primary_location.source.host_organization_name | Research Square (United States) |
| primary_location.source.host_organization_lineage | https://openalex.org/I4210096694 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.researchsquare.com/article/rs-1720857/latest.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.21203/rs.3.rs-1720857/v1 |
| publication_date | 2022-06-29 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W1980991473, https://openalex.org/W2058148991, https://openalex.org/W1971102564, https://openalex.org/W2074365347, https://openalex.org/W2921577782, https://openalex.org/W2029387902, https://openalex.org/W2338432197, https://openalex.org/W2043601400, https://openalex.org/W2096791516, https://openalex.org/W2115807273, https://openalex.org/W2101357408, https://openalex.org/W2041953582, https://openalex.org/W2098231283, https://openalex.org/W2076107693, https://openalex.org/W3098985301, https://openalex.org/W2098107333, https://openalex.org/W3023760086, https://openalex.org/W2136173440, https://openalex.org/W2793935388, https://openalex.org/W2800001480, https://openalex.org/W4301245661, https://openalex.org/W2973070265, https://openalex.org/W2143962878, https://openalex.org/W2308959237, https://openalex.org/W2169322959, https://openalex.org/W2789901287, https://openalex.org/W3023373652, https://openalex.org/W2100909778, https://openalex.org/W2515217061, https://openalex.org/W2891676927 |
| referenced_works_count | 30 |
| abstract_inverted_index.R | 212 |
| abstract_inverted_index.a | 6, 31, 42, 61, 80, 83, 199, 220, 277, 288 |
| abstract_inverted_index.To | 108, 158 |
| abstract_inverted_index.We | 181, 218 |
| abstract_inverted_index.Z) | 241 |
| abstract_inverted_index.an | 34, 114, 211 |
| abstract_inverted_index.as | 247 |
| abstract_inverted_index.at | 215 |
| abstract_inverted_index.by | 290 |
| abstract_inverted_index.in | 19, 30, 37, 73, 189, 210, 269 |
| abstract_inverted_index.is | 93, 208, 276 |
| abstract_inverted_index.of | 26, 41, 69, 98, 135, 156, 162, 170, 252 |
| abstract_inverted_index.on | 60, 121 |
| abstract_inverted_index.or | 67 |
| abstract_inverted_index.to | 11, 90, 238, 282 |
| abstract_inverted_index.we | 112, 164 |
| abstract_inverted_index.OWC | 175, 185, 275 |
| abstract_inverted_index.The | 24, 205 |
| abstract_inverted_index.and | 9, 104, 149, 198, 234, 242, 256 |
| abstract_inverted_index.are | 76, 102, 153 |
| abstract_inverted_index.can | 48 |
| abstract_inverted_index.for | 28, 82, 287 |
| abstract_inverted_index.its | 248 |
| abstract_inverted_index.sum | 134, 141 |
| abstract_inverted_index.the | 12, 39, 64, 150, 160, 253, 262, 283 |
| abstract_inverted_index.two | 235 |
| abstract_inverted_index.way | 25 |
| abstract_inverted_index.yet | 85 |
| abstract_inverted_index.(two | 231 |
| abstract_inverted_index.GWAS | 99, 125, 195, 202 |
| abstract_inverted_index.OWC, | 163, 265 |
| abstract_inverted_index.OWC. | 157 |
| abstract_inverted_index.Test | 147 |
| abstract_inverted_index.data | 75, 92, 101, 191, 197, 258 |
| abstract_inverted_index.four | 227 |
| abstract_inverted_index.from | 124 |
| abstract_inverted_index.gene | 32 |
| abstract_inverted_index.most | 270 |
| abstract_inverted_index.need | 81 |
| abstract_inverted_index.real | 74, 257 |
| abstract_inverted_index.role | 36 |
| abstract_inverted_index.show | 183 |
| abstract_inverted_index.test | 119, 138, 152, 224 |
| abstract_inverted_index.that | 174, 184, 225, 261, 274, 280 |
| abstract_inverted_index.tool | 279 |
| abstract_inverted_index.weak | 58 |
| abstract_inverted_index.when | 53 |
| abstract_inverted_index.with | 57 |
| abstract_inverted_index.(OWC) | 118 |
| abstract_inverted_index.These | 272 |
| abstract_inverted_index.based | 120 |
| abstract_inverted_index.boost | 49 |
| abstract_inverted_index.cases | 155 |
| abstract_inverted_index.data. | 126, 204 |
| abstract_inverted_index.model | 286 |
| abstract_inverted_index.novel | 221 |
| abstract_inverted_index.often | 94 |
| abstract_inverted_index.plays | 33 |
| abstract_inverted_index.power | 40 |
| abstract_inverted_index.score | 137, 151 |
| abstract_inverted_index.sizes | 66 |
| abstract_inverted_index.test, | 132, 264 |
| abstract_inverted_index.test. | 45 |
| abstract_inverted_index.tests | 4 |
| abstract_inverted_index.these | 110 |
| abstract_inverted_index.using | 193 |
| abstract_inverted_index.usual | 13 |
| abstract_inverted_index.while | 96 |
| abstract_inverted_index.Kernel | 145 |
| abstract_inverted_index.[SSU], | 139 |
| abstract_inverted_index.[WSS], | 143 |
| abstract_inverted_index.access | 89 |
| abstract_inverted_index.adapts | 281 |
| abstract_inverted_index.burden | 131 |
| abstract_inverted_index.cases. | 250 |
| abstract_inverted_index.causal | 70 |
| abstract_inverted_index.effect | 65 |
| abstract_inverted_index.freely | 105 |
| abstract_inverted_index.marker | 15 |
| abstract_inverted_index.method | 207 |
| abstract_inverted_index.normal | 239 |
| abstract_inverted_index.power, | 51 |
| abstract_inverted_index.robust | 86 |
| abstract_inverted_index.single | 14 |
| abstract_inverted_index.tests, | 17 |
| abstract_inverted_index.trait. | 62 |
| abstract_inverted_index.useful | 7, 278 |
| abstract_inverted_index.(GWAS). | 23 |
| abstract_inverted_index.Results | 169, 251 |
| abstract_inverted_index.SNP-set | 144 |
| abstract_inverted_index.Several | 127 |
| abstract_inverted_index.[SKAT], | 148 |
| abstract_inverted_index.disease | 289 |
| abstract_inverted_index.driving | 79 |
| abstract_inverted_index.effects | 59 |
| abstract_inverted_index.fasting | 200 |
| abstract_inverted_index.further | 182 |
| abstract_inverted_index.genetic | 55, 71 |
| abstract_inverted_index.glucose | 201 |
| abstract_inverted_index.methods | 129, 188, 246, 268 |
| abstract_inverted_index.package | 213 |
| abstract_inverted_index.perform | 165 |
| abstract_inverted_index.popular | 179, 245 |
| abstract_inverted_index.propose | 113, 219 |
| abstract_inverted_index.provide | 5 |
| abstract_inverted_index.resolve | 109 |
| abstract_inverted_index.schemes | 230 |
| abstract_inverted_index.several | 177, 244 |
| abstract_inverted_index.special | 154, 249 |
| abstract_inverted_index.squared | 136 |
| abstract_inverted_index.studies | 22, 172, 255 |
| abstract_inverted_index.summary | 100, 122, 196 |
| abstract_inverted_index.usually | 77 |
| abstract_inverted_index.weights | 47, 233, 236 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.However, | 63 |
| abstract_inverted_index.Results: | 107 |
| abstract_inverted_index.analyses | 192, 259 |
| abstract_inverted_index.boosting | 38 |
| abstract_inverted_index.constant | 232 |
| abstract_inverted_index.evaluate | 159 |
| abstract_inverted_index.existing | 178 |
| abstract_inverted_index.flexible | 84 |
| abstract_inverted_index.includes | 243 |
| abstract_inverted_index.limited, | 95 |
| abstract_inverted_index.methods. | 180 |
| abstract_inverted_index.proposed | 206, 263 |
| abstract_inverted_index.publicly | 103 |
| abstract_inverted_index.studies. | 168 |
| abstract_inverted_index.unknown, | 78 |
| abstract_inverted_index.variants | 29, 56, 72 |
| abstract_inverted_index.weighted | 116, 133, 140 |
| abstract_inverted_index.available | 214 |
| abstract_inverted_index.detecting | 54 |
| abstract_inverted_index.different | 228 |
| abstract_inverted_index.extensive | 166 |
| abstract_inverted_index.important | 35 |
| abstract_inverted_index.including | 130 |
| abstract_inverted_index.optimally | 115, 291 |
| abstract_inverted_index.statistic | 142, 240 |
| abstract_inverted_index.thousands | 97 |
| abstract_inverted_index.variants. | 293 |
| abstract_inverted_index.weighting | 27, 229, 292 |
| abstract_inverted_index.Gene-based | 2 |
| abstract_inverted_index.available. | 106 |
| abstract_inverted_index.biological | 285 |
| abstract_inverted_index.comparable | 267 |
| abstract_inverted_index.comparison | 187 |
| abstract_inverted_index.complement | 10 |
| abstract_inverted_index.directions | 68 |
| abstract_inverted_index.especially | 18, 52 |
| abstract_inverted_index.gene-based | 43, 222 |
| abstract_inverted_index.illustrate | 260 |
| abstract_inverted_index.real-world | 190 |
| abstract_inverted_index.scenarios. | 271 |
| abstract_inverted_index.simulation | 167, 171, 254 |
| abstract_inverted_index.statistics | 123 |
| abstract_inverted_index.underlying | 284 |
| abstract_inverted_index.Appropriate | 46 |
| abstract_inverted_index.Association | 146 |
| abstract_inverted_index.Background: | 1 |
| abstract_inverted_index.alternative | 8 |
| abstract_inverted_index.association | 3, 16, 21, 44, 223 |
| abstract_inverted_index.combination | 117 |
| abstract_inverted_index.demonstrate | 173, 273 |
| abstract_inverted_index.genome-wide | 20 |
| abstract_inverted_index.implemented | 209 |
| abstract_inverted_index.outperforms | 176, 186, 266 |
| abstract_inverted_index.performance | 161 |
| abstract_inverted_index.statistical | 50 |
| abstract_inverted_index.traditional | 128 |
| abstract_inverted_index.Conclusions: | 217 |
| abstract_inverted_index.Furthermore, | 88 |
| abstract_inverted_index.incorporates | 226 |
| abstract_inverted_index.limitations, | 111 |
| abstract_inverted_index.methodology. | 87 |
| abstract_inverted_index.proportional | 237 |
| abstract_inverted_index.meta-analysis | 203 |
| abstract_inverted_index.schizophrenia | 194 |
| abstract_inverted_index.individual-level | 91 |
| abstract_inverted_index.https://github.com/Jianjun-CN/R-package-for-OWC. | 216 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5101511802 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 6 |
| corresponding_institution_ids | https://openalex.org/I123534392 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/17 |
| sustainable_development_goals[0].score | 0.44999998807907104 |
| sustainable_development_goals[0].display_name | Partnerships for the goals |
| citation_normalized_percentile.value | 0.09033459 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |