A novel bacteriophage with broad host-range against Clostridioides difficile ribotype 078 elucidates the phage receptor Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.05.26.445907
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.05.26.445907
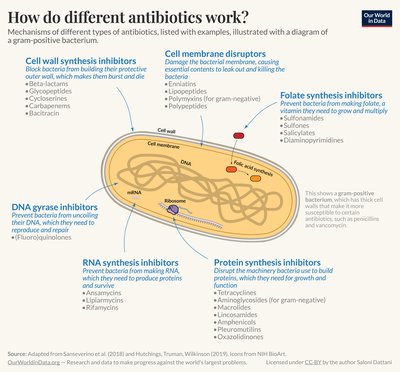
Bacteriophage represent a promising option for the treatment of Clostridioides difficile (formerly Clostridium difficile ) infection (CDI), which at present relies on conventional antibiotic therapy. The specificity of bacteriophages should prevent the dysbiosis of the colonic microbiota associated with the treatment of CDI with antibiotics. Whilst numerous phages have been isolated, none have been characterised with broad host-range activity towards PCR ribotype (RT) 078 C. difficile strains despite their considerable relevance to medicine and agriculture. In this study, we isolated four novel C. difficile Myoviruses: ΦCD08011, ΦCD418, ΦCD1801 and ΦCD2301. Their characterisation revealed that each was comparable with other C. difficile phages described in the literature, with the exception of ΦCD1801 which exhibited a broad host-range activity towards RT 078, infecting 15/16 (93.8%) of the clinical isolates tested. In order for wild-type phages to be exploited in the effective treatment of CDI, an optimal phage cocktail must be assembled that provides broad coverage against all C. difficile RTs. In an attempt to advance these efforts, we conducted a series of fundamental experiments that identified the C. difficile SlpA, the major constituent of the C. difficile surface-layer (S-layer), as the phage receptor. Thus, we demonstrated that ΦCD1801 could only bind to RT 012 or RT 027 strains in the presence of a plasmid-borne S-layer cassette corresponding to RT 078. Armed with this information, efforts should now be directed towards the isolation of phages with broad host-range activity against each of the fourteen described S-layer cassette types which could form the basis of an effective cocktail active against a wide range of C. difficile isolates. Importance Research into phage therapy has seen a resurgence in recent years owing to growing concerns regarding antimicrobial resistance. Phage research for potential therapy against Clostridium difficile infection (CDI) is in its infancy, where an optimal “one size fits all” phage cocktail is yet to be derived. The pursuit thus far, has aimed to find phages with the broadest possible host-range. Although, for C. difficile strains belonging to certain PCR ribotypes (RTs), in particular RT 078, phages with broad-host range activity are yet to be discovered. In this study, we isolate 4 novel Myoviruses including ΦCD1801, which exerts the broadest host-range activity towards RT 078 reported in the literature. Through the application of ΦCD1801 to robust binding assays, we elucidate SlpA as the phage receptor on the bacterial cell surface. Our finding suggests that an optimal “one size fits all” combinatorial phage cocktail, could theoretically comprise 14 phages, each targeting one of the 14 described S-layer cassettes of C. difficile .
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2021.05.26.445907
- https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdf
- OA Status
- green
- References
- 28
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3166483198
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3166483198Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2021.05.26.445907Digital Object Identifier
- Title
-
A novel bacteriophage with broad host-range against Clostridioides difficile ribotype 078 elucidates the phage receptorWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-05-29Full publication date if available
- Authors
-
Michaella Whittle, Terry W. Bilverstone, R. J. van Esveld, Arlen-Celina Lücke, Michelle M. Lister, Sarah A. Kuehne, Nigel P. MintonList of authors in order
- Landing page
-
https://doi.org/10.1101/2021.05.26.445907Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdfDirect OA link when available
- Concepts
-
Clostridium difficile, Bacteriophage, Microbiology, Clostridioides, Biology, Host (biology), Virology, Antibiotics, Genetics, Gene, Escherichia coliTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
28Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3166483198 |
|---|---|
| doi | https://doi.org/10.1101/2021.05.26.445907 |
| ids.doi | https://doi.org/10.1101/2021.05.26.445907 |
| ids.mag | 3166483198 |
| ids.openalex | https://openalex.org/W3166483198 |
| fwci | 0.0 |
| type | preprint |
| title | A novel bacteriophage with broad host-range against Clostridioides difficile ribotype 078 elucidates the phage receptor |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11288 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2725 |
| topics[0].subfield.display_name | Infectious Diseases |
| topics[0].display_name | Clostridium difficile and Clostridium perfringens research |
| topics[1].id | https://openalex.org/T10976 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9922999739646912 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2725 |
| topics[1].subfield.display_name | Infectious Diseases |
| topics[1].display_name | Viral gastroenteritis research and epidemiology |
| topics[2].id | https://openalex.org/T11048 |
| topics[2].field.id | https://openalex.org/fields/23 |
| topics[2].field.display_name | Environmental Science |
| topics[2].score | 0.9731000065803528 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2303 |
| topics[2].subfield.display_name | Ecology |
| topics[2].display_name | Bacteriophages and microbial interactions |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2994496256 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7456429600715637 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q27031090 |
| concepts[0].display_name | Clostridium difficile |
| concepts[1].id | https://openalex.org/C2776441376 |
| concepts[1].level | 4 |
| concepts[1].score | 0.7194594144821167 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q165028 |
| concepts[1].display_name | Bacteriophage |
| concepts[2].id | https://openalex.org/C89423630 |
| concepts[2].level | 1 |
| concepts[2].score | 0.7018507719039917 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[2].display_name | Microbiology |
| concepts[3].id | https://openalex.org/C2910067853 |
| concepts[3].level | 2 |
| concepts[3].score | 0.6327584981918335 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q27438828 |
| concepts[3].display_name | Clostridioides |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.5466998815536499 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C126831891 |
| concepts[5].level | 2 |
| concepts[5].score | 0.4910121262073517 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q221673 |
| concepts[5].display_name | Host (biology) |
| concepts[6].id | https://openalex.org/C159047783 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4441640079021454 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7215 |
| concepts[6].display_name | Virology |
| concepts[7].id | https://openalex.org/C501593827 |
| concepts[7].level | 2 |
| concepts[7].score | 0.39616331458091736 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q12187 |
| concepts[7].display_name | Antibiotics |
| concepts[8].id | https://openalex.org/C54355233 |
| concepts[8].level | 1 |
| concepts[8].score | 0.11847379803657532 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[8].display_name | Genetics |
| concepts[9].id | https://openalex.org/C104317684 |
| concepts[9].level | 2 |
| concepts[9].score | 0.10672476887702942 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[9].display_name | Gene |
| concepts[10].id | https://openalex.org/C547475151 |
| concepts[10].level | 3 |
| concepts[10].score | 0.09295594692230225 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q25419 |
| concepts[10].display_name | Escherichia coli |
| keywords[0].id | https://openalex.org/keywords/clostridium-difficile |
| keywords[0].score | 0.7456429600715637 |
| keywords[0].display_name | Clostridium difficile |
| keywords[1].id | https://openalex.org/keywords/bacteriophage |
| keywords[1].score | 0.7194594144821167 |
| keywords[1].display_name | Bacteriophage |
| keywords[2].id | https://openalex.org/keywords/microbiology |
| keywords[2].score | 0.7018507719039917 |
| keywords[2].display_name | Microbiology |
| keywords[3].id | https://openalex.org/keywords/clostridioides |
| keywords[3].score | 0.6327584981918335 |
| keywords[3].display_name | Clostridioides |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.5466998815536499 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/host |
| keywords[5].score | 0.4910121262073517 |
| keywords[5].display_name | Host (biology) |
| keywords[6].id | https://openalex.org/keywords/virology |
| keywords[6].score | 0.4441640079021454 |
| keywords[6].display_name | Virology |
| keywords[7].id | https://openalex.org/keywords/antibiotics |
| keywords[7].score | 0.39616331458091736 |
| keywords[7].display_name | Antibiotics |
| keywords[8].id | https://openalex.org/keywords/genetics |
| keywords[8].score | 0.11847379803657532 |
| keywords[8].display_name | Genetics |
| keywords[9].id | https://openalex.org/keywords/gene |
| keywords[9].score | 0.10672476887702942 |
| keywords[9].display_name | Gene |
| keywords[10].id | https://openalex.org/keywords/escherichia-coli |
| keywords[10].score | 0.09295594692230225 |
| keywords[10].display_name | Escherichia coli |
| language | en |
| locations[0].id | doi:10.1101/2021.05.26.445907 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2021.05.26.445907 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5007896636 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Michaella Whittle |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I1299528221, https://openalex.org/I142263535, https://openalex.org/I2799693246 |
| authorships[0].affiliations[0].raw_affiliation_string | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[0].institutions[0].id | https://openalex.org/I2799693246 |
| authorships[0].institutions[0].ror | https://ror.org/00cwqg982 |
| authorships[0].institutions[0].type | government |
| authorships[0].institutions[0].lineage | https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | Biotechnology and Biological Sciences Research Council |
| authorships[0].institutions[1].id | https://openalex.org/I1299528221 |
| authorships[0].institutions[1].ror | https://ror.org/0439y7842 |
| authorships[0].institutions[1].type | government |
| authorships[0].institutions[1].lineage | https://openalex.org/I1299528221, https://openalex.org/I4210087105 |
| authorships[0].institutions[1].country_code | GB |
| authorships[0].institutions[1].display_name | Engineering and Physical Sciences Research Council |
| authorships[0].institutions[2].id | https://openalex.org/I142263535 |
| authorships[0].institutions[2].ror | https://ror.org/01ee9ar58 |
| authorships[0].institutions[2].type | education |
| authorships[0].institutions[2].lineage | https://openalex.org/I142263535 |
| authorships[0].institutions[2].country_code | GB |
| authorships[0].institutions[2].display_name | University of Nottingham |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | M.J Whittle |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[1].author.id | https://openalex.org/A5015996661 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-7176-6917 |
| authorships[1].author.display_name | Terry W. Bilverstone |
| authorships[1].countries | GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I1299528221, https://openalex.org/I142263535, https://openalex.org/I2799693246 |
| authorships[1].affiliations[0].raw_affiliation_string | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[1].institutions[0].id | https://openalex.org/I2799693246 |
| authorships[1].institutions[0].ror | https://ror.org/00cwqg982 |
| authorships[1].institutions[0].type | government |
| authorships[1].institutions[0].lineage | https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | Biotechnology and Biological Sciences Research Council |
| authorships[1].institutions[1].id | https://openalex.org/I1299528221 |
| authorships[1].institutions[1].ror | https://ror.org/0439y7842 |
| authorships[1].institutions[1].type | government |
| authorships[1].institutions[1].lineage | https://openalex.org/I1299528221, https://openalex.org/I4210087105 |
| authorships[1].institutions[1].country_code | GB |
| authorships[1].institutions[1].display_name | Engineering and Physical Sciences Research Council |
| authorships[1].institutions[2].id | https://openalex.org/I142263535 |
| authorships[1].institutions[2].ror | https://ror.org/01ee9ar58 |
| authorships[1].institutions[2].type | education |
| authorships[1].institutions[2].lineage | https://openalex.org/I142263535 |
| authorships[1].institutions[2].country_code | GB |
| authorships[1].institutions[2].display_name | University of Nottingham |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | T.W Bilverstone |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[2].author.id | https://openalex.org/A5079559935 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | R. J. van Esveld |
| authorships[2].countries | NL |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I2800006345 |
| authorships[2].affiliations[0].raw_affiliation_string | Faculty of Medicine, Leiden University Medical Centre, the Netherlands |
| authorships[2].institutions[0].id | https://openalex.org/I2800006345 |
| authorships[2].institutions[0].ror | https://ror.org/05xvt9f17 |
| authorships[2].institutions[0].type | healthcare |
| authorships[2].institutions[0].lineage | https://openalex.org/I2800006345 |
| authorships[2].institutions[0].country_code | NL |
| authorships[2].institutions[0].display_name | Leiden University Medical Center |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | R.J van Esveld |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Faculty of Medicine, Leiden University Medical Centre, the Netherlands |
| authorships[3].author.id | https://openalex.org/A5012237887 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Arlen-Celina Lücke |
| authorships[3].countries | DE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I34809795 |
| authorships[3].affiliations[0].raw_affiliation_string | Hannover Medical School, Carl-Neuberg-Straße 1, 30625 Hannover, Germany |
| authorships[3].institutions[0].id | https://openalex.org/I34809795 |
| authorships[3].institutions[0].ror | https://ror.org/00f2yqf98 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I34809795 |
| authorships[3].institutions[0].country_code | DE |
| authorships[3].institutions[0].display_name | Medizinische Hochschule Hannover |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | A.C. Lücke |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Hannover Medical School, Carl-Neuberg-Straße 1, 30625 Hannover, Germany |
| authorships[4].author.id | https://openalex.org/A5065354157 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-5221-4102 |
| authorships[4].author.display_name | Michelle M. Lister |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I1299528221, https://openalex.org/I142263535, https://openalex.org/I2799693246 |
| authorships[4].affiliations[0].raw_affiliation_string | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I1334287468, https://openalex.org/I142263535, https://openalex.org/I4210146512 |
| authorships[4].affiliations[1].raw_affiliation_string | NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust and the University of Nottingham, Nottingham, NG7 RD, UK |
| authorships[4].affiliations[2].institution_ids | https://openalex.org/I1334287468, https://openalex.org/I4210137227 |
| authorships[4].affiliations[2].raw_affiliation_string | Department of Clinical Microbiology, Queen's Medical Centre, Nottingham University Hospitals NHS Trust, Nottingham, UK, UK |
| authorships[4].institutions[0].id | https://openalex.org/I2799693246 |
| authorships[4].institutions[0].ror | https://ror.org/00cwqg982 |
| authorships[4].institutions[0].type | government |
| authorships[4].institutions[0].lineage | https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | Biotechnology and Biological Sciences Research Council |
| authorships[4].institutions[1].id | https://openalex.org/I1299528221 |
| authorships[4].institutions[1].ror | https://ror.org/0439y7842 |
| authorships[4].institutions[1].type | government |
| authorships[4].institutions[1].lineage | https://openalex.org/I1299528221, https://openalex.org/I4210087105 |
| authorships[4].institutions[1].country_code | GB |
| authorships[4].institutions[1].display_name | Engineering and Physical Sciences Research Council |
| authorships[4].institutions[2].id | https://openalex.org/I4210146512 |
| authorships[4].institutions[2].ror | https://ror.org/046cr9566 |
| authorships[4].institutions[2].type | facility |
| authorships[4].institutions[2].lineage | https://openalex.org/I1334287468, https://openalex.org/I4210146512 |
| authorships[4].institutions[2].country_code | GB |
| authorships[4].institutions[2].display_name | Nottingham Biomedical Research Centre |
| authorships[4].institutions[3].id | https://openalex.org/I1334287468 |
| authorships[4].institutions[3].ror | https://ror.org/05y3qh794 |
| authorships[4].institutions[3].type | healthcare |
| authorships[4].institutions[3].lineage | https://openalex.org/I1334287468 |
| authorships[4].institutions[3].country_code | GB |
| authorships[4].institutions[3].display_name | Nottingham University Hospitals NHS Trust |
| authorships[4].institutions[4].id | https://openalex.org/I4210137227 |
| authorships[4].institutions[4].ror | https://ror.org/03ap6wx93 |
| authorships[4].institutions[4].type | healthcare |
| authorships[4].institutions[4].lineage | https://openalex.org/I1334287468, https://openalex.org/I4210137227 |
| authorships[4].institutions[4].country_code | GB |
| authorships[4].institutions[4].display_name | Queen's Medical Centre |
| authorships[4].institutions[5].id | https://openalex.org/I142263535 |
| authorships[4].institutions[5].ror | https://ror.org/01ee9ar58 |
| authorships[4].institutions[5].type | education |
| authorships[4].institutions[5].lineage | https://openalex.org/I142263535 |
| authorships[4].institutions[5].country_code | GB |
| authorships[4].institutions[5].display_name | University of Nottingham |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | M.M Lister |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK, Department of Clinical Microbiology, Queen's Medical Centre, Nottingham University Hospitals NHS Trust, Nottingham, UK, UK, NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust and the University of Nottingham, Nottingham, NG7 RD, UK |
| authorships[5].author.id | https://openalex.org/A5039213727 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-6790-8433 |
| authorships[5].author.display_name | Sarah A. Kuehne |
| authorships[5].countries | GB |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I1334287468, https://openalex.org/I142263535, https://openalex.org/I4210146512 |
| authorships[5].affiliations[0].raw_affiliation_string | NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust and the University of Nottingham, Nottingham, NG7 RD, UK |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I79619799 |
| authorships[5].affiliations[1].raw_affiliation_string | Oral Microbiology Group, School of Dentistry and Institute of Microbiology and Infection, College of Medical and Dental Sciences, The University of Birmingham, Birmingham, B5 7EG, UK |
| authorships[5].affiliations[2].institution_ids | https://openalex.org/I1299528221, https://openalex.org/I142263535, https://openalex.org/I2799693246 |
| authorships[5].affiliations[2].raw_affiliation_string | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[5].institutions[0].id | https://openalex.org/I2799693246 |
| authorships[5].institutions[0].ror | https://ror.org/00cwqg982 |
| authorships[5].institutions[0].type | government |
| authorships[5].institutions[0].lineage | https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[5].institutions[0].country_code | GB |
| authorships[5].institutions[0].display_name | Biotechnology and Biological Sciences Research Council |
| authorships[5].institutions[1].id | https://openalex.org/I1299528221 |
| authorships[5].institutions[1].ror | https://ror.org/0439y7842 |
| authorships[5].institutions[1].type | government |
| authorships[5].institutions[1].lineage | https://openalex.org/I1299528221, https://openalex.org/I4210087105 |
| authorships[5].institutions[1].country_code | GB |
| authorships[5].institutions[1].display_name | Engineering and Physical Sciences Research Council |
| authorships[5].institutions[2].id | https://openalex.org/I4210146512 |
| authorships[5].institutions[2].ror | https://ror.org/046cr9566 |
| authorships[5].institutions[2].type | facility |
| authorships[5].institutions[2].lineage | https://openalex.org/I1334287468, https://openalex.org/I4210146512 |
| authorships[5].institutions[2].country_code | GB |
| authorships[5].institutions[2].display_name | Nottingham Biomedical Research Centre |
| authorships[5].institutions[3].id | https://openalex.org/I1334287468 |
| authorships[5].institutions[3].ror | https://ror.org/05y3qh794 |
| authorships[5].institutions[3].type | healthcare |
| authorships[5].institutions[3].lineage | https://openalex.org/I1334287468 |
| authorships[5].institutions[3].country_code | GB |
| authorships[5].institutions[3].display_name | Nottingham University Hospitals NHS Trust |
| authorships[5].institutions[4].id | https://openalex.org/I79619799 |
| authorships[5].institutions[4].ror | https://ror.org/03angcq70 |
| authorships[5].institutions[4].type | education |
| authorships[5].institutions[4].lineage | https://openalex.org/I79619799 |
| authorships[5].institutions[4].country_code | GB |
| authorships[5].institutions[4].display_name | University of Birmingham |
| authorships[5].institutions[5].id | https://openalex.org/I142263535 |
| authorships[5].institutions[5].ror | https://ror.org/01ee9ar58 |
| authorships[5].institutions[5].type | education |
| authorships[5].institutions[5].lineage | https://openalex.org/I142263535 |
| authorships[5].institutions[5].country_code | GB |
| authorships[5].institutions[5].display_name | University of Nottingham |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | S.A Kuehne |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK, NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust and the University of Nottingham, Nottingham, NG7 RD, UK, Oral Microbiology Group, School of Dentistry and Institute of Microbiology and Infection, College of Medical and Dental Sciences, The University of Birmingham, Birmingham, B5 7EG, UK |
| authorships[6].author.id | https://openalex.org/A5044949409 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-9277-1261 |
| authorships[6].author.display_name | Nigel P. Minton |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1334287468, https://openalex.org/I142263535, https://openalex.org/I4210146512 |
| authorships[6].affiliations[0].raw_affiliation_string | NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust and the University of Nottingham, Nottingham, NG7 RD, UK |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I1299528221, https://openalex.org/I142263535, https://openalex.org/I2799693246 |
| authorships[6].affiliations[1].raw_affiliation_string | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK |
| authorships[6].institutions[0].id | https://openalex.org/I2799693246 |
| authorships[6].institutions[0].ror | https://ror.org/00cwqg982 |
| authorships[6].institutions[0].type | government |
| authorships[6].institutions[0].lineage | https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | Biotechnology and Biological Sciences Research Council |
| authorships[6].institutions[1].id | https://openalex.org/I1299528221 |
| authorships[6].institutions[1].ror | https://ror.org/0439y7842 |
| authorships[6].institutions[1].type | government |
| authorships[6].institutions[1].lineage | https://openalex.org/I1299528221, https://openalex.org/I4210087105 |
| authorships[6].institutions[1].country_code | GB |
| authorships[6].institutions[1].display_name | Engineering and Physical Sciences Research Council |
| authorships[6].institutions[2].id | https://openalex.org/I4210146512 |
| authorships[6].institutions[2].ror | https://ror.org/046cr9566 |
| authorships[6].institutions[2].type | facility |
| authorships[6].institutions[2].lineage | https://openalex.org/I1334287468, https://openalex.org/I4210146512 |
| authorships[6].institutions[2].country_code | GB |
| authorships[6].institutions[2].display_name | Nottingham Biomedical Research Centre |
| authorships[6].institutions[3].id | https://openalex.org/I1334287468 |
| authorships[6].institutions[3].ror | https://ror.org/05y3qh794 |
| authorships[6].institutions[3].type | healthcare |
| authorships[6].institutions[3].lineage | https://openalex.org/I1334287468 |
| authorships[6].institutions[3].country_code | GB |
| authorships[6].institutions[3].display_name | Nottingham University Hospitals NHS Trust |
| authorships[6].institutions[4].id | https://openalex.org/I142263535 |
| authorships[6].institutions[4].ror | https://ror.org/01ee9ar58 |
| authorships[6].institutions[4].type | education |
| authorships[6].institutions[4].lineage | https://openalex.org/I142263535 |
| authorships[6].institutions[4].country_code | GB |
| authorships[6].institutions[4].display_name | University of Nottingham |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | N.P Minton |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Clostridia Research Group, BBSRC/EPSRC Synthetic Biology Research Centre (SBRC), School of Life Sciences, Biodiscovery Institute, The University of Nottingham, Nottingham, NG7 2RD, UK, NIHR Nottingham Biomedical Research Centre, Nottingham University Hospitals NHS Trust and the University of Nottingham, Nottingham, NG7 RD, UK |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | A novel bacteriophage with broad host-range against Clostridioides difficile ribotype 078 elucidates the phage receptor |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11288 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2725 |
| primary_topic.subfield.display_name | Infectious Diseases |
| primary_topic.display_name | Clostridium difficile and Clostridium perfringens research |
| related_works | https://openalex.org/W4283034770, https://openalex.org/W3154202355, https://openalex.org/W2995316057, https://openalex.org/W3166585310, https://openalex.org/W3205563078, https://openalex.org/W4383956697, https://openalex.org/W4382895319, https://openalex.org/W2946545977, https://openalex.org/W4403724532, https://openalex.org/W3128876904 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2021.05.26.445907 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2021.05.26.445907 |
| primary_location.id | doi:10.1101/2021.05.26.445907 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2021/05/29/2021.05.26.445907.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2021.05.26.445907 |
| publication_date | 2021-05-29 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2464112039, https://openalex.org/W2071054182, https://openalex.org/W2087083032, https://openalex.org/W2043965252, https://openalex.org/W2139607253, https://openalex.org/W2515956436, https://openalex.org/W2807560728, https://openalex.org/W2253087149, https://openalex.org/W2751560363, https://openalex.org/W2883298628, https://openalex.org/W2258870953, https://openalex.org/W2137726039, https://openalex.org/W2049355215, https://openalex.org/W2158524196, https://openalex.org/W2552844454, https://openalex.org/W1996423252, https://openalex.org/W2127385500, https://openalex.org/W2345791363, https://openalex.org/W2431034466, https://openalex.org/W2147218692, https://openalex.org/W2119169151, https://openalex.org/W1977241950, https://openalex.org/W1576302805, https://openalex.org/W4236872823, https://openalex.org/W2170488656, https://openalex.org/W2090591880, https://openalex.org/W1980238990, https://openalex.org/W3123255347 |
| referenced_works_count | 28 |
| abstract_inverted_index.) | 15 |
| abstract_inverted_index.. | 424 |
| abstract_inverted_index.4 | 355 |
| abstract_inverted_index.a | 3, 114, 168, 211, 257, 271 |
| abstract_inverted_index.14 | 410, 417 |
| abstract_inverted_index.C. | 65, 83, 100, 156, 176, 184, 261, 327, 422 |
| abstract_inverted_index.In | 76, 129, 159, 350 |
| abstract_inverted_index.RT | 119, 201, 204, 217, 338, 367 |
| abstract_inverted_index.an | 143, 160, 252, 298, 398 |
| abstract_inverted_index.as | 188, 385 |
| abstract_inverted_index.at | 19 |
| abstract_inverted_index.be | 135, 148, 226, 309, 348 |
| abstract_inverted_index.in | 104, 137, 207, 273, 294, 336, 370 |
| abstract_inverted_index.is | 293, 306 |
| abstract_inverted_index.of | 9, 28, 34, 42, 110, 124, 141, 170, 182, 210, 231, 239, 251, 260, 376, 415, 421 |
| abstract_inverted_index.on | 22, 389 |
| abstract_inverted_index.or | 203 |
| abstract_inverted_index.to | 72, 134, 162, 200, 216, 277, 308, 317, 331, 347, 378 |
| abstract_inverted_index.we | 79, 166, 193, 353, 382 |
| abstract_inverted_index.012 | 202 |
| abstract_inverted_index.027 | 205 |
| abstract_inverted_index.078 | 64, 368 |
| abstract_inverted_index.CDI | 43 |
| abstract_inverted_index.Our | 394 |
| abstract_inverted_index.PCR | 61, 333 |
| abstract_inverted_index.The | 26, 311 |
| abstract_inverted_index.all | 155 |
| abstract_inverted_index.and | 74, 89 |
| abstract_inverted_index.are | 345 |
| abstract_inverted_index.for | 6, 131, 285, 326 |
| abstract_inverted_index.has | 269, 315 |
| abstract_inverted_index.its | 295 |
| abstract_inverted_index.now | 225 |
| abstract_inverted_index.one | 414 |
| abstract_inverted_index.the | 7, 32, 35, 40, 105, 108, 125, 138, 175, 179, 183, 189, 208, 229, 240, 249, 321, 362, 371, 374, 386, 390, 416 |
| abstract_inverted_index.was | 96 |
| abstract_inverted_index.yet | 307, 346 |
| abstract_inverted_index.(RT) | 63 |
| abstract_inverted_index.078, | 120, 339 |
| abstract_inverted_index.078. | 218 |
| abstract_inverted_index.CDI, | 142 |
| abstract_inverted_index.RTs. | 158 |
| abstract_inverted_index.SlpA | 384 |
| abstract_inverted_index.been | 50, 54 |
| abstract_inverted_index.bind | 199 |
| abstract_inverted_index.cell | 392 |
| abstract_inverted_index.each | 95, 238, 412 |
| abstract_inverted_index.far, | 314 |
| abstract_inverted_index.find | 318 |
| abstract_inverted_index.fits | 302, 402 |
| abstract_inverted_index.form | 248 |
| abstract_inverted_index.four | 81 |
| abstract_inverted_index.have | 49, 53 |
| abstract_inverted_index.into | 266 |
| abstract_inverted_index.must | 147 |
| abstract_inverted_index.none | 52 |
| abstract_inverted_index.only | 198 |
| abstract_inverted_index.seen | 270 |
| abstract_inverted_index.size | 301, 401 |
| abstract_inverted_index.that | 94, 150, 173, 195, 397 |
| abstract_inverted_index.this | 77, 221, 351 |
| abstract_inverted_index.thus | 313 |
| abstract_inverted_index.wide | 258 |
| abstract_inverted_index.with | 39, 44, 56, 98, 107, 220, 233, 320, 341 |
| abstract_inverted_index.(CDI) | 292 |
| abstract_inverted_index.15/16 | 122 |
| abstract_inverted_index.Armed | 219 |
| abstract_inverted_index.Phage | 283 |
| abstract_inverted_index.SlpA, | 178 |
| abstract_inverted_index.Their | 91 |
| abstract_inverted_index.Thus, | 192 |
| abstract_inverted_index.aimed | 316 |
| abstract_inverted_index.basis | 250 |
| abstract_inverted_index.broad | 57, 115, 152, 234 |
| abstract_inverted_index.could | 197, 247, 407 |
| abstract_inverted_index.major | 180 |
| abstract_inverted_index.novel | 82, 356 |
| abstract_inverted_index.order | 130 |
| abstract_inverted_index.other | 99 |
| abstract_inverted_index.owing | 276 |
| abstract_inverted_index.phage | 145, 190, 267, 304, 387, 405 |
| abstract_inverted_index.range | 259, 343 |
| abstract_inverted_index.their | 69 |
| abstract_inverted_index.these | 164 |
| abstract_inverted_index.types | 245 |
| abstract_inverted_index.where | 297 |
| abstract_inverted_index.which | 18, 112, 246, 360 |
| abstract_inverted_index.years | 275 |
| abstract_inverted_index.(CDI), | 17 |
| abstract_inverted_index.(RTs), | 335 |
| abstract_inverted_index.Whilst | 46 |
| abstract_inverted_index.active | 255 |
| abstract_inverted_index.all” | 303, 403 |
| abstract_inverted_index.exerts | 361 |
| abstract_inverted_index.option | 5 |
| abstract_inverted_index.phages | 48, 102, 133, 232, 319, 340 |
| abstract_inverted_index.recent | 274 |
| abstract_inverted_index.relies | 21 |
| abstract_inverted_index.robust | 379 |
| abstract_inverted_index.series | 169 |
| abstract_inverted_index.should | 30, 224 |
| abstract_inverted_index.study, | 78, 352 |
| abstract_inverted_index.“one | 300, 400 |
| abstract_inverted_index.(93.8%) | 123 |
| abstract_inverted_index.S-layer | 213, 243, 419 |
| abstract_inverted_index.Through | 373 |
| abstract_inverted_index.advance | 163 |
| abstract_inverted_index.against | 154, 237, 256, 288 |
| abstract_inverted_index.assays, | 381 |
| abstract_inverted_index.attempt | 161 |
| abstract_inverted_index.binding | 380 |
| abstract_inverted_index.certain | 332 |
| abstract_inverted_index.colonic | 36 |
| abstract_inverted_index.despite | 68 |
| abstract_inverted_index.efforts | 223 |
| abstract_inverted_index.finding | 395 |
| abstract_inverted_index.growing | 278 |
| abstract_inverted_index.isolate | 354 |
| abstract_inverted_index.optimal | 144, 299, 399 |
| abstract_inverted_index.phages, | 411 |
| abstract_inverted_index.present | 20 |
| abstract_inverted_index.prevent | 31 |
| abstract_inverted_index.pursuit | 312 |
| abstract_inverted_index.strains | 67, 206, 329 |
| abstract_inverted_index.tested. | 128 |
| abstract_inverted_index.therapy | 268, 287 |
| abstract_inverted_index.towards | 60, 118, 228, 366 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Research | 265 |
| abstract_inverted_index.activity | 59, 117, 236, 344, 365 |
| abstract_inverted_index.broadest | 322, 363 |
| abstract_inverted_index.cassette | 214, 244 |
| abstract_inverted_index.clinical | 126 |
| abstract_inverted_index.cocktail | 146, 254, 305 |
| abstract_inverted_index.comprise | 409 |
| abstract_inverted_index.concerns | 279 |
| abstract_inverted_index.coverage | 153 |
| abstract_inverted_index.derived. | 310 |
| abstract_inverted_index.directed | 227 |
| abstract_inverted_index.efforts, | 165 |
| abstract_inverted_index.fourteen | 241 |
| abstract_inverted_index.infancy, | 296 |
| abstract_inverted_index.isolated | 80 |
| abstract_inverted_index.isolates | 127 |
| abstract_inverted_index.medicine | 73 |
| abstract_inverted_index.numerous | 47 |
| abstract_inverted_index.possible | 323 |
| abstract_inverted_index.presence | 209 |
| abstract_inverted_index.provides | 151 |
| abstract_inverted_index.receptor | 388 |
| abstract_inverted_index.reported | 369 |
| abstract_inverted_index.research | 284 |
| abstract_inverted_index.revealed | 93 |
| abstract_inverted_index.ribotype | 62 |
| abstract_inverted_index.suggests | 396 |
| abstract_inverted_index.surface. | 393 |
| abstract_inverted_index.therapy. | 25 |
| abstract_inverted_index.ΦCD1801 | 88, 111, 196, 377 |
| abstract_inverted_index.ΦCD418, | 87 |
| abstract_inverted_index.(formerly | 12 |
| abstract_inverted_index.Although, | 325 |
| abstract_inverted_index.assembled | 149 |
| abstract_inverted_index.bacterial | 391 |
| abstract_inverted_index.belonging | 330 |
| abstract_inverted_index.cassettes | 420 |
| abstract_inverted_index.cocktail, | 406 |
| abstract_inverted_index.conducted | 167 |
| abstract_inverted_index.described | 103, 242, 418 |
| abstract_inverted_index.difficile | 11, 14, 66, 84, 101, 157, 177, 185, 262, 290, 328, 423 |
| abstract_inverted_index.dysbiosis | 33 |
| abstract_inverted_index.effective | 139, 253 |
| abstract_inverted_index.elucidate | 383 |
| abstract_inverted_index.exception | 109 |
| abstract_inverted_index.exhibited | 113 |
| abstract_inverted_index.exploited | 136 |
| abstract_inverted_index.including | 358 |
| abstract_inverted_index.infecting | 121 |
| abstract_inverted_index.infection | 16, 291 |
| abstract_inverted_index.isolated, | 51 |
| abstract_inverted_index.isolates. | 263 |
| abstract_inverted_index.isolation | 230 |
| abstract_inverted_index.potential | 286 |
| abstract_inverted_index.promising | 4 |
| abstract_inverted_index.receptor. | 191 |
| abstract_inverted_index.regarding | 280 |
| abstract_inverted_index.relevance | 71 |
| abstract_inverted_index.represent | 2 |
| abstract_inverted_index.ribotypes | 334 |
| abstract_inverted_index.targeting | 413 |
| abstract_inverted_index.treatment | 8, 41, 140 |
| abstract_inverted_index.wild-type | 132 |
| abstract_inverted_index.ΦCD1801, | 359 |
| abstract_inverted_index.ΦCD2301. | 90 |
| abstract_inverted_index.(S-layer), | 187 |
| abstract_inverted_index.Importance | 264 |
| abstract_inverted_index.Myoviruses | 357 |
| abstract_inverted_index.antibiotic | 24 |
| abstract_inverted_index.associated | 38 |
| abstract_inverted_index.broad-host | 342 |
| abstract_inverted_index.comparable | 97 |
| abstract_inverted_index.host-range | 58, 116, 235, 364 |
| abstract_inverted_index.identified | 174 |
| abstract_inverted_index.microbiota | 37 |
| abstract_inverted_index.particular | 337 |
| abstract_inverted_index.resurgence | 272 |
| abstract_inverted_index.ΦCD08011, | 86 |
| abstract_inverted_index.Clostridium | 13, 289 |
| abstract_inverted_index.Myoviruses: | 85 |
| abstract_inverted_index.application | 375 |
| abstract_inverted_index.constituent | 181 |
| abstract_inverted_index.discovered. | 349 |
| abstract_inverted_index.experiments | 172 |
| abstract_inverted_index.fundamental | 171 |
| abstract_inverted_index.host-range. | 324 |
| abstract_inverted_index.literature, | 106 |
| abstract_inverted_index.literature. | 372 |
| abstract_inverted_index.resistance. | 282 |
| abstract_inverted_index.specificity | 27 |
| abstract_inverted_index.agriculture. | 75 |
| abstract_inverted_index.antibiotics. | 45 |
| abstract_inverted_index.considerable | 70 |
| abstract_inverted_index.conventional | 23 |
| abstract_inverted_index.demonstrated | 194 |
| abstract_inverted_index.information, | 222 |
| abstract_inverted_index.Bacteriophage | 1 |
| abstract_inverted_index.antimicrobial | 281 |
| abstract_inverted_index.characterised | 55 |
| abstract_inverted_index.combinatorial | 404 |
| abstract_inverted_index.corresponding | 215 |
| abstract_inverted_index.plasmid-borne | 212 |
| abstract_inverted_index.surface-layer | 186 |
| abstract_inverted_index.theoretically | 408 |
| abstract_inverted_index.Clostridioides | 10 |
| abstract_inverted_index.bacteriophages | 29 |
| abstract_inverted_index.characterisation | 92 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5044949409 |
| countries_distinct_count | 3 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I1299528221, https://openalex.org/I1334287468, https://openalex.org/I142263535, https://openalex.org/I2799693246, https://openalex.org/I4210146512 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/2 |
| sustainable_development_goals[0].score | 0.7400000095367432 |
| sustainable_development_goals[0].display_name | Zero hunger |
| citation_normalized_percentile.value | 0.06070757 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |