A novel computational methodology for GWAS multi-locus analysis based on graph theory and machine learning Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.10.22.21265388
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.10.22.21265388
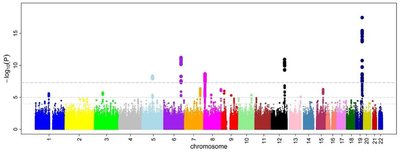
Background Current form of genome-wide association studies (GWAS) is inadequate to accurately explain the genetics of complex traits due to the lack of sufficient statistical power. It explores each variant individually, but current studies show that multiple variants with varying effect sizes actually act in a concerted way to develop a complex disease. To address this issue, we have developed an algorithmic framework that can effectively solve the multi-locus problem in GWAS with a very high level of confidence. Our methodology consists of three novel algorithms based on graph theory and machine learning. It identifies a set of highly discriminating variants that are stable and robust with little (if any) spuriousness. Consequently, likely these variants should be able to interpret missing heritability of a convoluted disease as an entity. Results To demonstrate the efficacy of our proposed algorithms, we have considered astigmatism case-control GWAS dataset. Astigmatism is a common eye condition that causes blurred vision because of an error in the shape of the cornea. The cause of astigmatism is not entirely known but a sizable inheritability is assumed. Clinical studies show that developmental disorders (such as, autism) and astigmatism co-occur in a statistically significant number of individuals. By performing classical GWAS analysis, we didn’t find any genome-wide statistically significant variants. Conversely, we have identified a set of stable, robust, and highly predictive variants that can together explain the genetics of astigmatism. We have performed a set of biological enrichment analyses based on gene ontology (GO) terms, disease ontology (DO) terms, biological pathways, network of pathways, and so forth to manifest the accuracy and novelty of our findings. Conclusions Rigorous experimental evaluations show that our proposed methodology can solve GWAS multi-locus problem effectively and efficiently. It can identify signals from the GWAS dataset having small number of samples with a high level of accuracy. We believe that the proposed methodology based on graph theory and machine learning is the most comprehensive one compared to any other machine learning based tools in this domain.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2021.10.22.21265388
- https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdf
- OA Status
- green
- References
- 68
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3210271793
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3210271793Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2021.10.22.21265388Digital Object Identifier
- Title
-
A novel computational methodology for GWAS multi-locus analysis based on graph theory and machine learningWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-10-26Full publication date if available
- Authors
-
Subrata Saha, Himanshu Narayan Singh, Ahmed Soliman, Sanguthevar RajasekaranList of authors in order
- Landing page
-
https://doi.org/10.1101/2021.10.22.21265388Publisher landing page
- PDF URL
-
https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdfDirect OA link when available
- Concepts
-
Genome-wide association study, Locus (genetics), Genetic association, Heritability, Missing heritability problem, Computer science, Artificial intelligence, Computational biology, Biology, Machine learning, Genetics, Genetic variants, Gene, Genotype, Single-nucleotide polymorphismTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
68Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3210271793 |
|---|---|
| doi | https://doi.org/10.1101/2021.10.22.21265388 |
| ids.doi | https://doi.org/10.1101/2021.10.22.21265388 |
| ids.mag | 3210271793 |
| ids.openalex | https://openalex.org/W3210271793 |
| fwci | 0.0 |
| type | preprint |
| title | A novel computational methodology for GWAS multi-locus analysis based on graph theory and machine learning |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10261 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9980000257492065 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic Associations and Epidemiology |
| topics[1].id | https://openalex.org/T11213 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9965999722480774 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genomic variations and chromosomal abnormalities |
| topics[2].id | https://openalex.org/T11642 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9846000075340271 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genomics and Rare Diseases |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.8480985164642334 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C84597430 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5218942761421204 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q106227 |
| concepts[1].display_name | Locus (genetics) |
| concepts[2].id | https://openalex.org/C186413461 |
| concepts[2].level | 5 |
| concepts[2].score | 0.5040287375450134 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[2].display_name | Genetic association |
| concepts[3].id | https://openalex.org/C161890455 |
| concepts[3].level | 2 |
| concepts[3].score | 0.4632173180580139 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1503548 |
| concepts[3].display_name | Heritability |
| concepts[4].id | https://openalex.org/C144621757 |
| concepts[4].level | 5 |
| concepts[4].score | 0.45754191279411316 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q3083720 |
| concepts[4].display_name | Missing heritability problem |
| concepts[5].id | https://openalex.org/C41008148 |
| concepts[5].level | 0 |
| concepts[5].score | 0.44367432594299316 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[5].display_name | Computer science |
| concepts[6].id | https://openalex.org/C154945302 |
| concepts[6].level | 1 |
| concepts[6].score | 0.397771418094635 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[6].display_name | Artificial intelligence |
| concepts[7].id | https://openalex.org/C70721500 |
| concepts[7].level | 1 |
| concepts[7].score | 0.39178016781806946 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[7].display_name | Computational biology |
| concepts[8].id | https://openalex.org/C86803240 |
| concepts[8].level | 0 |
| concepts[8].score | 0.3662372827529907 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[8].display_name | Biology |
| concepts[9].id | https://openalex.org/C119857082 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3554930090904236 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[9].display_name | Machine learning |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.2927858233451843 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C2993967602 |
| concepts[11].level | 4 |
| concepts[11].score | 0.21165764331817627 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q5532932 |
| concepts[11].display_name | Genetic variants |
| concepts[12].id | https://openalex.org/C104317684 |
| concepts[12].level | 2 |
| concepts[12].score | 0.1901651918888092 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[12].display_name | Gene |
| concepts[13].id | https://openalex.org/C135763542 |
| concepts[13].level | 3 |
| concepts[13].score | 0.10394507646560669 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[13].display_name | Genotype |
| concepts[14].id | https://openalex.org/C153209595 |
| concepts[14].level | 4 |
| concepts[14].score | 0.09906718134880066 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[14].display_name | Single-nucleotide polymorphism |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.8480985164642334 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/locus |
| keywords[1].score | 0.5218942761421204 |
| keywords[1].display_name | Locus (genetics) |
| keywords[2].id | https://openalex.org/keywords/genetic-association |
| keywords[2].score | 0.5040287375450134 |
| keywords[2].display_name | Genetic association |
| keywords[3].id | https://openalex.org/keywords/heritability |
| keywords[3].score | 0.4632173180580139 |
| keywords[3].display_name | Heritability |
| keywords[4].id | https://openalex.org/keywords/missing-heritability-problem |
| keywords[4].score | 0.45754191279411316 |
| keywords[4].display_name | Missing heritability problem |
| keywords[5].id | https://openalex.org/keywords/computer-science |
| keywords[5].score | 0.44367432594299316 |
| keywords[5].display_name | Computer science |
| keywords[6].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[6].score | 0.397771418094635 |
| keywords[6].display_name | Artificial intelligence |
| keywords[7].id | https://openalex.org/keywords/computational-biology |
| keywords[7].score | 0.39178016781806946 |
| keywords[7].display_name | Computational biology |
| keywords[8].id | https://openalex.org/keywords/biology |
| keywords[8].score | 0.3662372827529907 |
| keywords[8].display_name | Biology |
| keywords[9].id | https://openalex.org/keywords/machine-learning |
| keywords[9].score | 0.3554930090904236 |
| keywords[9].display_name | Machine learning |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.2927858233451843 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/genetic-variants |
| keywords[11].score | 0.21165764331817627 |
| keywords[11].display_name | Genetic variants |
| keywords[12].id | https://openalex.org/keywords/gene |
| keywords[12].score | 0.1901651918888092 |
| keywords[12].display_name | Gene |
| keywords[13].id | https://openalex.org/keywords/genotype |
| keywords[13].score | 0.10394507646560669 |
| keywords[13].display_name | Genotype |
| keywords[14].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[14].score | 0.09906718134880066 |
| keywords[14].display_name | Single-nucleotide polymorphism |
| language | en |
| locations[0].id | doi:10.1101/2021.10.22.21265388 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2021.10.22.21265388 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5100725452 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-2991-0481 |
| authorships[0].author.display_name | Subrata Saha |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I2799503643 |
| authorships[0].affiliations[0].raw_affiliation_string | Irving Medical Center, Columbia University New York, NY 10032, US |
| authorships[0].institutions[0].id | https://openalex.org/I2799503643 |
| authorships[0].institutions[0].ror | https://ror.org/01esghr10 |
| authorships[0].institutions[0].type | healthcare |
| authorships[0].institutions[0].lineage | https://openalex.org/I2799503643 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Columbia University Irving Medical Center |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Subrata Saha |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Irving Medical Center, Columbia University New York, NY 10032, US |
| authorships[1].author.id | https://openalex.org/A5071080377 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-4111-5165 |
| authorships[1].author.display_name | Himanshu Narayan Singh |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I2799503643 |
| authorships[1].affiliations[0].raw_affiliation_string | Irving Medical Center, Columbia University New York, NY 10032, US |
| authorships[1].institutions[0].id | https://openalex.org/I2799503643 |
| authorships[1].institutions[0].ror | https://ror.org/01esghr10 |
| authorships[1].institutions[0].type | healthcare |
| authorships[1].institutions[0].lineage | https://openalex.org/I2799503643 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Columbia University Irving Medical Center |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Himanshu Narayan Singh |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Irving Medical Center, Columbia University New York, NY 10032, US |
| authorships[2].author.id | https://openalex.org/A5079036569 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-4339-1385 |
| authorships[2].author.display_name | Ahmed Soliman |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I140172145 |
| authorships[2].affiliations[0].raw_affiliation_string | University of Connecticut, Department of Computer Science and Engineering, Storrs, CT 06269, US |
| authorships[2].institutions[0].id | https://openalex.org/I140172145 |
| authorships[2].institutions[0].ror | https://ror.org/02der9h97 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I140172145 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of Connecticut |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Ahmed Soliman |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | University of Connecticut, Department of Computer Science and Engineering, Storrs, CT 06269, US |
| authorships[3].author.id | https://openalex.org/A5034177039 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Sanguthevar Rajasekaran |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I140172145 |
| authorships[3].affiliations[0].raw_affiliation_string | University of Connecticut, Department of Computer Science and Engineering, Storrs, CT 06269, US |
| authorships[3].institutions[0].id | https://openalex.org/I140172145 |
| authorships[3].institutions[0].ror | https://ror.org/02der9h97 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I140172145 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of Connecticut |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Sanguthevar Rajasekaran |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | University of Connecticut, Department of Computer Science and Engineering, Storrs, CT 06269, US |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | A novel computational methodology for GWAS multi-locus analysis based on graph theory and machine learning |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10261 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9980000257492065 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic Associations and Epidemiology |
| related_works | https://openalex.org/W159064268, https://openalex.org/W2136285728, https://openalex.org/W2111896105, https://openalex.org/W2170506381, https://openalex.org/W1987620105, https://openalex.org/W2102533238, https://openalex.org/W1975304626, https://openalex.org/W1847429134, https://openalex.org/W2144000849, https://openalex.org/W2067245483 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2021.10.22.21265388 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2021.10.22.21265388 |
| primary_location.id | doi:10.1101/2021.10.22.21265388 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.medrxiv.org/content/medrxiv/early/2021/10/26/2021.10.22.21265388.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2021.10.22.21265388 |
| publication_date | 2021-10-26 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2169294950, https://openalex.org/W2125858955, https://openalex.org/W2992152394, https://openalex.org/W3185306500, https://openalex.org/W2016511593, https://openalex.org/W2558493806, https://openalex.org/W2122227569, https://openalex.org/W2108380304, https://openalex.org/W2959750635, https://openalex.org/W2948009788, https://openalex.org/W2091239497, https://openalex.org/W2006712471, https://openalex.org/W2087347434, https://openalex.org/W4239510810, https://openalex.org/W2108995755, https://openalex.org/W2035720976, https://openalex.org/W4205247356, https://openalex.org/W2143238378, https://openalex.org/W2109372707, https://openalex.org/W2905452503, https://openalex.org/W2521967673, https://openalex.org/W2051674484, https://openalex.org/W2111326065, https://openalex.org/W2915985519, https://openalex.org/W2102619694, https://openalex.org/W2141820415, https://openalex.org/W2161633633, https://openalex.org/W2794134966, https://openalex.org/W3175752499, https://openalex.org/W2151014102, https://openalex.org/W2417483443, https://openalex.org/W2018884294, https://openalex.org/W2018844166, https://openalex.org/W2314123452, https://openalex.org/W2537679995, https://openalex.org/W2273769940, https://openalex.org/W2002122073, https://openalex.org/W2403402366, https://openalex.org/W2809346511, https://openalex.org/W1964287112, https://openalex.org/W1998594238, https://openalex.org/W2804295544, https://openalex.org/W2059496726, https://openalex.org/W1988011192, https://openalex.org/W1935300532, https://openalex.org/W2905115911, https://openalex.org/W2145744991, https://openalex.org/W2620588197, https://openalex.org/W2089238445, https://openalex.org/W2013645919, https://openalex.org/W2752705695, https://openalex.org/W3020961160, https://openalex.org/W3185128241, https://openalex.org/W1979540749, https://openalex.org/W1537717727, https://openalex.org/W1996429511, https://openalex.org/W2310152563, https://openalex.org/W2996699344, https://openalex.org/W4251663120, https://openalex.org/W2006576003, https://openalex.org/W2795080268, https://openalex.org/W3125405901, https://openalex.org/W2119821739, https://openalex.org/W3091619938, https://openalex.org/W2127640118, https://openalex.org/W2235816398, https://openalex.org/W2070895433, https://openalex.org/W2127553917 |
| referenced_works_count | 68 |
| abstract_inverted_index.a | 46, 51, 74, 96, 124, 148, 175, 193, 216, 236, 300 |
| abstract_inverted_index.By | 199 |
| abstract_inverted_index.It | 27, 94, 286 |
| abstract_inverted_index.To | 54, 131 |
| abstract_inverted_index.We | 233, 305 |
| abstract_inverted_index.an | 61, 128, 158 |
| abstract_inverted_index.as | 127 |
| abstract_inverted_index.be | 117 |
| abstract_inverted_index.in | 45, 71, 160, 192, 331 |
| abstract_inverted_index.is | 9, 147, 170, 178, 318 |
| abstract_inverted_index.of | 4, 16, 23, 78, 83, 98, 123, 135, 157, 163, 168, 197, 218, 231, 238, 255, 266, 297, 303 |
| abstract_inverted_index.on | 88, 243, 312 |
| abstract_inverted_index.so | 258 |
| abstract_inverted_index.to | 11, 20, 49, 119, 260, 324 |
| abstract_inverted_index.we | 58, 139, 204, 213 |
| abstract_inverted_index.(if | 109 |
| abstract_inverted_index.Our | 80 |
| abstract_inverted_index.The | 166 |
| abstract_inverted_index.act | 44 |
| abstract_inverted_index.and | 91, 105, 189, 221, 257, 264, 284, 315 |
| abstract_inverted_index.any | 207, 325 |
| abstract_inverted_index.are | 103 |
| abstract_inverted_index.as, | 187 |
| abstract_inverted_index.but | 32, 174 |
| abstract_inverted_index.can | 65, 226, 278, 287 |
| abstract_inverted_index.due | 19 |
| abstract_inverted_index.eye | 150 |
| abstract_inverted_index.not | 171 |
| abstract_inverted_index.one | 322 |
| abstract_inverted_index.our | 136, 267, 275 |
| abstract_inverted_index.set | 97, 217, 237 |
| abstract_inverted_index.the | 14, 21, 68, 133, 161, 164, 229, 262, 291, 308, 319 |
| abstract_inverted_index.way | 48 |
| abstract_inverted_index.(DO) | 250 |
| abstract_inverted_index.(GO) | 246 |
| abstract_inverted_index.GWAS | 72, 144, 202, 280, 292 |
| abstract_inverted_index.able | 118 |
| abstract_inverted_index.any) | 110 |
| abstract_inverted_index.each | 29 |
| abstract_inverted_index.find | 206 |
| abstract_inverted_index.form | 3 |
| abstract_inverted_index.from | 290 |
| abstract_inverted_index.gene | 244 |
| abstract_inverted_index.have | 59, 140, 214, 234 |
| abstract_inverted_index.high | 76, 301 |
| abstract_inverted_index.lack | 22 |
| abstract_inverted_index.most | 320 |
| abstract_inverted_index.show | 35, 182, 273 |
| abstract_inverted_index.that | 36, 64, 102, 152, 183, 225, 274, 307 |
| abstract_inverted_index.this | 56, 332 |
| abstract_inverted_index.very | 75 |
| abstract_inverted_index.with | 39, 73, 107, 299 |
| abstract_inverted_index.(such | 186 |
| abstract_inverted_index.based | 87, 242, 311, 329 |
| abstract_inverted_index.cause | 167 |
| abstract_inverted_index.error | 159 |
| abstract_inverted_index.forth | 259 |
| abstract_inverted_index.graph | 89, 313 |
| abstract_inverted_index.known | 173 |
| abstract_inverted_index.level | 77, 302 |
| abstract_inverted_index.novel | 85 |
| abstract_inverted_index.other | 326 |
| abstract_inverted_index.shape | 162 |
| abstract_inverted_index.sizes | 42 |
| abstract_inverted_index.small | 295 |
| abstract_inverted_index.solve | 67, 279 |
| abstract_inverted_index.these | 114 |
| abstract_inverted_index.three | 84 |
| abstract_inverted_index.tools | 330 |
| abstract_inverted_index.(GWAS) | 8 |
| abstract_inverted_index.causes | 153 |
| abstract_inverted_index.common | 149 |
| abstract_inverted_index.effect | 41 |
| abstract_inverted_index.having | 294 |
| abstract_inverted_index.highly | 99, 222 |
| abstract_inverted_index.issue, | 57 |
| abstract_inverted_index.likely | 113 |
| abstract_inverted_index.little | 108 |
| abstract_inverted_index.number | 196, 296 |
| abstract_inverted_index.power. | 26 |
| abstract_inverted_index.robust | 106 |
| abstract_inverted_index.should | 116 |
| abstract_inverted_index.stable | 104 |
| abstract_inverted_index.terms, | 247, 251 |
| abstract_inverted_index.theory | 90, 314 |
| abstract_inverted_index.traits | 18 |
| abstract_inverted_index.vision | 155 |
| abstract_inverted_index.Current | 2 |
| abstract_inverted_index.Results | 130 |
| abstract_inverted_index.address | 55 |
| abstract_inverted_index.autism) | 188 |
| abstract_inverted_index.because | 156 |
| abstract_inverted_index.believe | 306 |
| abstract_inverted_index.blurred | 154 |
| abstract_inverted_index.complex | 17, 52 |
| abstract_inverted_index.cornea. | 165 |
| abstract_inverted_index.current | 33 |
| abstract_inverted_index.dataset | 293 |
| abstract_inverted_index.develop | 50 |
| abstract_inverted_index.disease | 126, 248 |
| abstract_inverted_index.domain. | 333 |
| abstract_inverted_index.entity. | 129 |
| abstract_inverted_index.explain | 13, 228 |
| abstract_inverted_index.machine | 92, 316, 327 |
| abstract_inverted_index.missing | 121 |
| abstract_inverted_index.network | 254 |
| abstract_inverted_index.novelty | 265 |
| abstract_inverted_index.problem | 70, 282 |
| abstract_inverted_index.robust, | 220 |
| abstract_inverted_index.samples | 298 |
| abstract_inverted_index.signals | 289 |
| abstract_inverted_index.sizable | 176 |
| abstract_inverted_index.stable, | 219 |
| abstract_inverted_index.studies | 7, 34, 181 |
| abstract_inverted_index.variant | 30 |
| abstract_inverted_index.varying | 40 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Clinical | 180 |
| abstract_inverted_index.Rigorous | 270 |
| abstract_inverted_index.accuracy | 263 |
| abstract_inverted_index.actually | 43 |
| abstract_inverted_index.analyses | 241 |
| abstract_inverted_index.assumed. | 179 |
| abstract_inverted_index.co-occur | 191 |
| abstract_inverted_index.compared | 323 |
| abstract_inverted_index.consists | 82 |
| abstract_inverted_index.dataset. | 145 |
| abstract_inverted_index.didn’t | 205 |
| abstract_inverted_index.disease. | 53 |
| abstract_inverted_index.efficacy | 134 |
| abstract_inverted_index.entirely | 172 |
| abstract_inverted_index.explores | 28 |
| abstract_inverted_index.genetics | 15, 230 |
| abstract_inverted_index.identify | 288 |
| abstract_inverted_index.learning | 317, 328 |
| abstract_inverted_index.manifest | 261 |
| abstract_inverted_index.multiple | 37 |
| abstract_inverted_index.ontology | 245, 249 |
| abstract_inverted_index.proposed | 137, 276, 309 |
| abstract_inverted_index.together | 227 |
| abstract_inverted_index.variants | 38, 101, 115, 224 |
| abstract_inverted_index.accuracy. | 304 |
| abstract_inverted_index.analysis, | 203 |
| abstract_inverted_index.classical | 201 |
| abstract_inverted_index.concerted | 47 |
| abstract_inverted_index.condition | 151 |
| abstract_inverted_index.developed | 60 |
| abstract_inverted_index.disorders | 185 |
| abstract_inverted_index.findings. | 268 |
| abstract_inverted_index.framework | 63 |
| abstract_inverted_index.interpret | 120 |
| abstract_inverted_index.learning. | 93 |
| abstract_inverted_index.pathways, | 253, 256 |
| abstract_inverted_index.performed | 235 |
| abstract_inverted_index.variants. | 211 |
| abstract_inverted_index.Background | 1 |
| abstract_inverted_index.accurately | 12 |
| abstract_inverted_index.algorithms | 86 |
| abstract_inverted_index.biological | 239, 252 |
| abstract_inverted_index.considered | 141 |
| abstract_inverted_index.convoluted | 125 |
| abstract_inverted_index.enrichment | 240 |
| abstract_inverted_index.identified | 215 |
| abstract_inverted_index.identifies | 95 |
| abstract_inverted_index.inadequate | 10 |
| abstract_inverted_index.performing | 200 |
| abstract_inverted_index.predictive | 223 |
| abstract_inverted_index.sufficient | 24 |
| abstract_inverted_index.Astigmatism | 146 |
| abstract_inverted_index.Conclusions | 269 |
| abstract_inverted_index.Conversely, | 212 |
| abstract_inverted_index.algorithmic | 62 |
| abstract_inverted_index.algorithms, | 138 |
| abstract_inverted_index.association | 6 |
| abstract_inverted_index.astigmatism | 142, 169, 190 |
| abstract_inverted_index.confidence. | 79 |
| abstract_inverted_index.demonstrate | 132 |
| abstract_inverted_index.effectively | 66, 283 |
| abstract_inverted_index.evaluations | 272 |
| abstract_inverted_index.genome-wide | 5, 208 |
| abstract_inverted_index.methodology | 81, 277, 310 |
| abstract_inverted_index.multi-locus | 69, 281 |
| abstract_inverted_index.significant | 195, 210 |
| abstract_inverted_index.statistical | 25 |
| abstract_inverted_index.astigmatism. | 232 |
| abstract_inverted_index.case-control | 143 |
| abstract_inverted_index.efficiently. | 285 |
| abstract_inverted_index.experimental | 271 |
| abstract_inverted_index.heritability | 122 |
| abstract_inverted_index.individuals. | 198 |
| abstract_inverted_index.Consequently, | 112 |
| abstract_inverted_index.comprehensive | 321 |
| abstract_inverted_index.developmental | 184 |
| abstract_inverted_index.individually, | 31 |
| abstract_inverted_index.spuriousness. | 111 |
| abstract_inverted_index.statistically | 194, 209 |
| abstract_inverted_index.discriminating | 100 |
| abstract_inverted_index.inheritability | 177 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5034177039 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 4 |
| corresponding_institution_ids | https://openalex.org/I140172145 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/10 |
| sustainable_development_goals[0].score | 0.6200000047683716 |
| sustainable_development_goals[0].display_name | Reduced inequalities |
| citation_normalized_percentile.value | 0.15076677 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |