A SET domain-containing protein and HCF-1 maintain transgenerational epigenetic memory Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.03.25.645221
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.03.25.645221
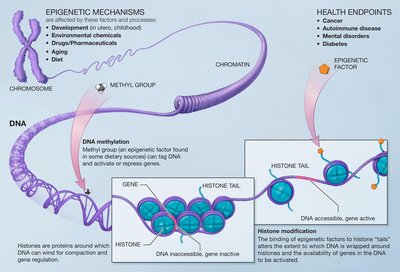
Epigenetic information can be inherited through a process known as transgenerational epigenetic inheritance (TEI). TEI can be initiated and maintained by small RNAs and histone modifications. The latter includes histone methylation, deposited by proteins with methyltransferase activity, including SET domain-containing proteins. Other SET domain-containing proteins with no catalytic methyltransferase activity also adopt roles in chromatin and gene expression regulation. Here, we describe SET-24, a SET domain-containing protein in Caenorhabditis elegans that belongs to a family of catalytically inactive SET proteins. SET-24 localises to germline nuclei and is required for germline immortality. Additionally, the inheritance of small RNA-driven epigenetic silencing is compromised in set-24 mutants. Using quantitative proteomics and yeast two-hybrid assays, we found that SET-24 interacts with Host Cell Factor 1 (HCF-1), a protein involved in epigenetic regulation and associated with known chromatin remodelling complexes, like COMPASS, which deposits H3K4me3. In set-24 mutants, hundreds of genes display increased H3K4me3 levels at their transcription start sites. While these changes are not matched at the transcriptional level, small RNA production is disrupted in approximately one fifth of those genes, which are normally targeted by small RNA pathways. We propose that SET-24 is a factor required to maintain epigenetic memory in the germline by maintaining a chromatin environment permissible to small RNA biogenesis over generations.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2025.03.25.645221
- https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdf
- OA Status
- green
- Cited By
- 1
- References
- 134
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4408889995
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4408889995Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2025.03.25.645221Digital Object Identifier
- Title
-
A SET domain-containing protein and HCF-1 maintain transgenerational epigenetic memoryWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-03-27Full publication date if available
- Authors
-
Chenming Zeng, Giulia Furlan, Miguel Vasconcelos Almeida, Juan C. Rueda-Silva, Jonathan Price, Jonas Mars, Pedro Rebelo‐Guiomar, Meng Huang, Shouhong Guang, Falk Butter, Eric A. MiskaList of authors in order
- Landing page
-
https://doi.org/10.1101/2025.03.25.645221Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdfDirect OA link when available
- Concepts
-
Transgenerational epigenetics, Epigenetics, Set (abstract data type), Domain (mathematical analysis), Computer science, Biology, Genetics, Computational biology, Mathematics, Gene, Programming language, Mathematical analysisTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
134Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4408889995 |
|---|---|
| doi | https://doi.org/10.1101/2025.03.25.645221 |
| ids.doi | https://doi.org/10.1101/2025.03.25.645221 |
| ids.openalex | https://openalex.org/W4408889995 |
| fwci | 2.68294463 |
| type | preprint |
| title | A SET domain-containing protein and HCF-1 maintain transgenerational epigenetic memory |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10269 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9520000219345093 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Epigenetics and DNA Methylation |
| topics[1].id | https://openalex.org/T11772 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9182000160217285 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genetics and Neurodevelopmental Disorders |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C184151982 |
| concepts[0].level | 4 |
| concepts[0].score | 0.9371392726898193 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q7834236 |
| concepts[0].display_name | Transgenerational epigenetics |
| concepts[1].id | https://openalex.org/C41091548 |
| concepts[1].level | 3 |
| concepts[1].score | 0.7677148580551147 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q26939 |
| concepts[1].display_name | Epigenetics |
| concepts[2].id | https://openalex.org/C177264268 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6131555438041687 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q1514741 |
| concepts[2].display_name | Set (abstract data type) |
| concepts[3].id | https://openalex.org/C36503486 |
| concepts[3].level | 2 |
| concepts[3].score | 0.6010978817939758 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q11235244 |
| concepts[3].display_name | Domain (mathematical analysis) |
| concepts[4].id | https://openalex.org/C41008148 |
| concepts[4].level | 0 |
| concepts[4].score | 0.39878571033477783 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[4].display_name | Computer science |
| concepts[5].id | https://openalex.org/C86803240 |
| concepts[5].level | 0 |
| concepts[5].score | 0.38511922955513 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[5].display_name | Biology |
| concepts[6].id | https://openalex.org/C54355233 |
| concepts[6].level | 1 |
| concepts[6].score | 0.35710301995277405 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[6].display_name | Genetics |
| concepts[7].id | https://openalex.org/C70721500 |
| concepts[7].level | 1 |
| concepts[7].score | 0.3290282189846039 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[7].display_name | Computational biology |
| concepts[8].id | https://openalex.org/C33923547 |
| concepts[8].level | 0 |
| concepts[8].score | 0.19204890727996826 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[8].display_name | Mathematics |
| concepts[9].id | https://openalex.org/C104317684 |
| concepts[9].level | 2 |
| concepts[9].score | 0.15514180064201355 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[9].display_name | Gene |
| concepts[10].id | https://openalex.org/C199360897 |
| concepts[10].level | 1 |
| concepts[10].score | 0.05344671010971069 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[10].display_name | Programming language |
| concepts[11].id | https://openalex.org/C134306372 |
| concepts[11].level | 1 |
| concepts[11].score | 0.0 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7754 |
| concepts[11].display_name | Mathematical analysis |
| keywords[0].id | https://openalex.org/keywords/transgenerational-epigenetics |
| keywords[0].score | 0.9371392726898193 |
| keywords[0].display_name | Transgenerational epigenetics |
| keywords[1].id | https://openalex.org/keywords/epigenetics |
| keywords[1].score | 0.7677148580551147 |
| keywords[1].display_name | Epigenetics |
| keywords[2].id | https://openalex.org/keywords/set |
| keywords[2].score | 0.6131555438041687 |
| keywords[2].display_name | Set (abstract data type) |
| keywords[3].id | https://openalex.org/keywords/domain |
| keywords[3].score | 0.6010978817939758 |
| keywords[3].display_name | Domain (mathematical analysis) |
| keywords[4].id | https://openalex.org/keywords/computer-science |
| keywords[4].score | 0.39878571033477783 |
| keywords[4].display_name | Computer science |
| keywords[5].id | https://openalex.org/keywords/biology |
| keywords[5].score | 0.38511922955513 |
| keywords[5].display_name | Biology |
| keywords[6].id | https://openalex.org/keywords/genetics |
| keywords[6].score | 0.35710301995277405 |
| keywords[6].display_name | Genetics |
| keywords[7].id | https://openalex.org/keywords/computational-biology |
| keywords[7].score | 0.3290282189846039 |
| keywords[7].display_name | Computational biology |
| keywords[8].id | https://openalex.org/keywords/mathematics |
| keywords[8].score | 0.19204890727996826 |
| keywords[8].display_name | Mathematics |
| keywords[9].id | https://openalex.org/keywords/gene |
| keywords[9].score | 0.15514180064201355 |
| keywords[9].display_name | Gene |
| keywords[10].id | https://openalex.org/keywords/programming-language |
| keywords[10].score | 0.05344671010971069 |
| keywords[10].display_name | Programming language |
| language | en |
| locations[0].id | doi:10.1101/2025.03.25.645221 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2025.03.25.645221 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5079504305 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Chenming Zeng |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Chenming Zeng |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5050888688 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-6134-6564 |
| authorships[1].author.display_name | Giulia Furlan |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Giulia Furlan |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5060430028 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-4816-3111 |
| authorships[2].author.display_name | Miguel Vasconcelos Almeida |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Miguel Vasconcelos Almeida |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5116818210 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Juan C. Rueda-Silva |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Juan C. Rueda-Silva |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5058827251 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-6554-5667 |
| authorships[4].author.display_name | Jonathan Price |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Jonathan Price |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5078857974 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-5904-2235 |
| authorships[5].author.display_name | Jonas Mars |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Jonas Mars |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5003461757 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-5060-7519 |
| authorships[6].author.display_name | Pedro Rebelo‐Guiomar |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Pedro Rebelo-Guiomar |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5070265823 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-6493-8223 |
| authorships[7].author.display_name | Meng Huang |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Meng Huang |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5013457687 |
| authorships[8].author.orcid | https://orcid.org/0000-0001-7700-9634 |
| authorships[8].author.display_name | Shouhong Guang |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Shouhong Guang |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5026716265 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-7197-7279 |
| authorships[9].author.display_name | Falk Butter |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Falk Butter |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5019197621 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-4450-576X |
| authorships[10].author.display_name | Eric A. Miska |
| authorships[10].author_position | last |
| authorships[10].raw_author_name | Eric A. Miska |
| authorships[10].is_corresponding | False |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | A SET domain-containing protein and HCF-1 maintain transgenerational epigenetic memory |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10269 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9520000219345093 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Epigenetics and DNA Methylation |
| related_works | https://openalex.org/W2035122032, https://openalex.org/W2971982268, https://openalex.org/W2773237536, https://openalex.org/W2738218039, https://openalex.org/W2790572587, https://openalex.org/W1991175997, https://openalex.org/W1491524348, https://openalex.org/W2330598502, https://openalex.org/W1886199067, https://openalex.org/W1577435787 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2025.03.25.645221 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2025.03.25.645221 |
| primary_location.id | doi:10.1101/2025.03.25.645221 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2025/03/27/2025.03.25.645221.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2025.03.25.645221 |
| publication_date | 2025-03-27 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W1647075334, https://openalex.org/W1979220516, https://openalex.org/W2091591117, https://openalex.org/W2080248723, https://openalex.org/W2038720758, https://openalex.org/W1885681284, https://openalex.org/W4297092696, https://openalex.org/W3195854644, https://openalex.org/W2018295108, https://openalex.org/W2122984483, https://openalex.org/W2922361588, https://openalex.org/W3049685990, https://openalex.org/W2154222068, https://openalex.org/W2560233932, https://openalex.org/W1861196385, https://openalex.org/W2095282684, https://openalex.org/W2804348858, https://openalex.org/W2901793276, https://openalex.org/W3173246325, https://openalex.org/W4404091329, https://openalex.org/W2786896132, https://openalex.org/W2601640824, https://openalex.org/W2588395757, https://openalex.org/W2790591579, https://openalex.org/W2032640887, https://openalex.org/W2617239062, https://openalex.org/W3034374923, https://openalex.org/W2089759992, https://openalex.org/W1585010992, https://openalex.org/W2619990190, https://openalex.org/W2804476437, https://openalex.org/W2783495821, https://openalex.org/W3003974482, https://openalex.org/W4388852446, https://openalex.org/W2807591486, https://openalex.org/W1575147957, https://openalex.org/W2979183639, https://openalex.org/W2088035403, https://openalex.org/W4297265151, https://openalex.org/W1960425032, https://openalex.org/W2802973814, https://openalex.org/W4387765899, https://openalex.org/W2586743691, https://openalex.org/W1997173542, https://openalex.org/W2546928489, https://openalex.org/W2903704117, https://openalex.org/W1963842907, https://openalex.org/W3160723104, https://openalex.org/W4394581743, https://openalex.org/W3028900815, https://openalex.org/W2018039678, https://openalex.org/W3017535517, https://openalex.org/W4283327445, https://openalex.org/W2111217101, https://openalex.org/W2787719063, https://openalex.org/W2788865254, https://openalex.org/W2015423672, https://openalex.org/W2095491514, https://openalex.org/W2063071041, https://openalex.org/W2091863961, https://openalex.org/W4392817832, https://openalex.org/W1987523347, https://openalex.org/W2003053220, https://openalex.org/W2030823261, https://openalex.org/W2979783981, https://openalex.org/W2120478300, https://openalex.org/W4399909411, https://openalex.org/W2022936129, https://openalex.org/W2287479634, https://openalex.org/W3090472322, https://openalex.org/W2170843897, https://openalex.org/W2885271663, https://openalex.org/W1969263950, https://openalex.org/W4406089136, https://openalex.org/W1969575940, https://openalex.org/W2921654140, https://openalex.org/W2007126009, https://openalex.org/W2000685943, https://openalex.org/W2019111498, https://openalex.org/W3028807786, https://openalex.org/W2050889043, https://openalex.org/W2951350919, https://openalex.org/W2899256508, https://openalex.org/W3081887529, https://openalex.org/W3080871630, https://openalex.org/W3138188786, https://openalex.org/W2020639485, https://openalex.org/W2157917165, https://openalex.org/W2606412267, https://openalex.org/W3084234242, https://openalex.org/W3095583226, https://openalex.org/W4308616064, https://openalex.org/W2160378127, https://openalex.org/W2559577985, https://openalex.org/W3005235420, https://openalex.org/W2951464304, https://openalex.org/W4396721167, https://openalex.org/W4375858802, https://openalex.org/W2734356283, https://openalex.org/W2976082486, https://openalex.org/W2114570899, https://openalex.org/W1991366673, https://openalex.org/W1986656413, https://openalex.org/W2080752012, https://openalex.org/W2949236108, https://openalex.org/W2103441770, https://openalex.org/W2108234281, https://openalex.org/W2171808845, https://openalex.org/W2179438025, https://openalex.org/W6911590169, https://openalex.org/W3035965352, https://openalex.org/W3003257820, https://openalex.org/W2011301426, https://openalex.org/W2131271579, https://openalex.org/W2592811885, https://openalex.org/W2952363993, https://openalex.org/W2036897871, https://openalex.org/W2169456326, https://openalex.org/W4406690981, https://openalex.org/W2138207763, https://openalex.org/W4393444553, https://openalex.org/W2990427812, https://openalex.org/W2128599926, https://openalex.org/W2131478115, https://openalex.org/W2122914905, https://openalex.org/W2803862617, https://openalex.org/W2100336754, https://openalex.org/W2124134646, https://openalex.org/W1970679013, https://openalex.org/W1995048848, https://openalex.org/W2129785061, https://openalex.org/W2341539131, https://openalex.org/W2112029506, https://openalex.org/W2411397641 |
| referenced_works_count | 134 |
| abstract_inverted_index.1 | 121 |
| abstract_inverted_index.a | 7, 64, 74, 123, 191, 203 |
| abstract_inverted_index.In | 141 |
| abstract_inverted_index.We | 186 |
| abstract_inverted_index.as | 10 |
| abstract_inverted_index.at | 151, 162 |
| abstract_inverted_index.be | 4, 17 |
| abstract_inverted_index.by | 21, 33, 182, 201 |
| abstract_inverted_index.in | 54, 68, 102, 126, 171, 198 |
| abstract_inverted_index.is | 87, 100, 169, 190 |
| abstract_inverted_index.no | 47 |
| abstract_inverted_index.of | 76, 95, 145, 175 |
| abstract_inverted_index.to | 73, 83, 194, 207 |
| abstract_inverted_index.we | 61, 112 |
| abstract_inverted_index.RNA | 167, 184, 209 |
| abstract_inverted_index.SET | 39, 43, 65, 79 |
| abstract_inverted_index.TEI | 15 |
| abstract_inverted_index.The | 27 |
| abstract_inverted_index.and | 19, 24, 56, 86, 108, 129 |
| abstract_inverted_index.are | 159, 179 |
| abstract_inverted_index.can | 3, 16 |
| abstract_inverted_index.for | 89 |
| abstract_inverted_index.not | 160 |
| abstract_inverted_index.one | 173 |
| abstract_inverted_index.the | 93, 163, 199 |
| abstract_inverted_index.Cell | 119 |
| abstract_inverted_index.Host | 118 |
| abstract_inverted_index.RNAs | 23 |
| abstract_inverted_index.also | 51 |
| abstract_inverted_index.gene | 57 |
| abstract_inverted_index.like | 136 |
| abstract_inverted_index.over | 211 |
| abstract_inverted_index.that | 71, 114, 188 |
| abstract_inverted_index.with | 35, 46, 117, 131 |
| abstract_inverted_index.Here, | 60 |
| abstract_inverted_index.Other | 42 |
| abstract_inverted_index.Using | 105 |
| abstract_inverted_index.While | 156 |
| abstract_inverted_index.adopt | 52 |
| abstract_inverted_index.fifth | 174 |
| abstract_inverted_index.found | 113 |
| abstract_inverted_index.genes | 146 |
| abstract_inverted_index.known | 9, 132 |
| abstract_inverted_index.roles | 53 |
| abstract_inverted_index.small | 22, 96, 166, 183, 208 |
| abstract_inverted_index.start | 154 |
| abstract_inverted_index.their | 152 |
| abstract_inverted_index.these | 157 |
| abstract_inverted_index.those | 176 |
| abstract_inverted_index.which | 138, 178 |
| abstract_inverted_index.yeast | 109 |
| abstract_inverted_index.(TEI). | 14 |
| abstract_inverted_index.Factor | 120 |
| abstract_inverted_index.SET-24 | 81, 115, 189 |
| abstract_inverted_index.factor | 192 |
| abstract_inverted_index.family | 75 |
| abstract_inverted_index.genes, | 177 |
| abstract_inverted_index.latter | 28 |
| abstract_inverted_index.level, | 165 |
| abstract_inverted_index.levels | 150 |
| abstract_inverted_index.memory | 197 |
| abstract_inverted_index.nuclei | 85 |
| abstract_inverted_index.set-24 | 103, 142 |
| abstract_inverted_index.sites. | 155 |
| abstract_inverted_index.H3K4me3 | 149 |
| abstract_inverted_index.SET-24, | 63 |
| abstract_inverted_index.assays, | 111 |
| abstract_inverted_index.belongs | 72 |
| abstract_inverted_index.changes | 158 |
| abstract_inverted_index.display | 147 |
| abstract_inverted_index.elegans | 70 |
| abstract_inverted_index.histone | 25, 30 |
| abstract_inverted_index.matched | 161 |
| abstract_inverted_index.process | 8 |
| abstract_inverted_index.propose | 187 |
| abstract_inverted_index.protein | 67, 124 |
| abstract_inverted_index.through | 6 |
| abstract_inverted_index.(HCF-1), | 122 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.COMPASS, | 137 |
| abstract_inverted_index.H3K4me3. | 140 |
| abstract_inverted_index.activity | 50 |
| abstract_inverted_index.deposits | 139 |
| abstract_inverted_index.describe | 62 |
| abstract_inverted_index.germline | 84, 90, 200 |
| abstract_inverted_index.hundreds | 144 |
| abstract_inverted_index.inactive | 78 |
| abstract_inverted_index.includes | 29 |
| abstract_inverted_index.involved | 125 |
| abstract_inverted_index.maintain | 195 |
| abstract_inverted_index.mutants, | 143 |
| abstract_inverted_index.mutants. | 104 |
| abstract_inverted_index.normally | 180 |
| abstract_inverted_index.proteins | 34, 45 |
| abstract_inverted_index.required | 88, 193 |
| abstract_inverted_index.targeted | 181 |
| abstract_inverted_index.activity, | 37 |
| abstract_inverted_index.catalytic | 48 |
| abstract_inverted_index.chromatin | 55, 133, 204 |
| abstract_inverted_index.deposited | 32 |
| abstract_inverted_index.disrupted | 170 |
| abstract_inverted_index.including | 38 |
| abstract_inverted_index.increased | 148 |
| abstract_inverted_index.inherited | 5 |
| abstract_inverted_index.initiated | 18 |
| abstract_inverted_index.interacts | 116 |
| abstract_inverted_index.localises | 82 |
| abstract_inverted_index.pathways. | 185 |
| abstract_inverted_index.proteins. | 41, 80 |
| abstract_inverted_index.silencing | 99 |
| abstract_inverted_index.Epigenetic | 1 |
| abstract_inverted_index.RNA-driven | 97 |
| abstract_inverted_index.associated | 130 |
| abstract_inverted_index.biogenesis | 210 |
| abstract_inverted_index.complexes, | 135 |
| abstract_inverted_index.epigenetic | 12, 98, 127, 196 |
| abstract_inverted_index.expression | 58 |
| abstract_inverted_index.maintained | 20 |
| abstract_inverted_index.production | 168 |
| abstract_inverted_index.proteomics | 107 |
| abstract_inverted_index.regulation | 128 |
| abstract_inverted_index.two-hybrid | 110 |
| abstract_inverted_index.compromised | 101 |
| abstract_inverted_index.environment | 205 |
| abstract_inverted_index.information | 2 |
| abstract_inverted_index.inheritance | 13, 94 |
| abstract_inverted_index.maintaining | 202 |
| abstract_inverted_index.permissible | 206 |
| abstract_inverted_index.regulation. | 59 |
| abstract_inverted_index.remodelling | 134 |
| abstract_inverted_index.generations. | 212 |
| abstract_inverted_index.immortality. | 91 |
| abstract_inverted_index.methylation, | 31 |
| abstract_inverted_index.quantitative | 106 |
| abstract_inverted_index.Additionally, | 92 |
| abstract_inverted_index.approximately | 172 |
| abstract_inverted_index.catalytically | 77 |
| abstract_inverted_index.transcription | 153 |
| abstract_inverted_index.Caenorhabditis | 69 |
| abstract_inverted_index.modifications. | 26 |
| abstract_inverted_index.transcriptional | 164 |
| abstract_inverted_index.domain-containing | 40, 44, 66 |
| abstract_inverted_index.methyltransferase | 36, 49 |
| abstract_inverted_index.transgenerational | 11 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 11 |
| citation_normalized_percentile.value | 0.78074706 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |