A Simple Way to Incorporate Target Structural Information in Molecular Generative Models Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.02.17.529000
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.02.17.529000
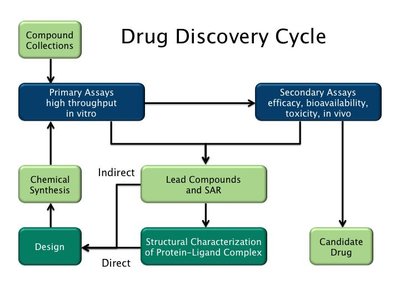
Deep learning generative models are now being applied in various fields including drug discovery. In this work, we propose a novel approach to include target 3D structural information in molecular generative models for structure-based drug design. The method combines a message-passing neural network model that predicts docking scores with a generative neural network model as its reward function to navigate the chemical space searching for molecules that bind favorably with a specific target. A key feature of the method is the construction of target-specific molecular sets for training, designed to overcome potential transferability issues of surrogate docking models through a two-round training process. Consequently, this enables accurate guided exploration of the chemical space without reliance on the collection of prior knowledge about active and inactive compounds for the specific target. Tests on eight target proteins showed a 100-fold increase in hit generation compared to conventional docking calculations, and the ability to generate molecules similar to approved drugs or known active ligands for specific targets without prior knowledge. This method provides a general and highly efficient solution for structure-based molecular generation.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.02.17.529000
- https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdf
- OA Status
- green
- References
- 57
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4321444361
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4321444361Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.02.17.529000Digital Object Identifier
- Title
-
A Simple Way to Incorporate Target Structural Information in Molecular Generative ModelsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-02-21Full publication date if available
- Authors
-
Wenyi Zhang, Kaiyue Zhang, Jing HuangList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.02.17.529000Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdfDirect OA link when available
- Concepts
-
Computer science, Generative grammar, Artificial intelligence, Artificial neural network, Generative model, Machine learning, Chemical space, Docking (animal), Transferability, Training set, Drug discovery, Drug target, Chemistry, Bioinformatics, Biology, Biochemistry, Medicine, Nursing, LogitTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
57Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4321444361 |
|---|---|
| doi | https://doi.org/10.1101/2023.02.17.529000 |
| ids.doi | https://doi.org/10.1101/2023.02.17.529000 |
| ids.openalex | https://openalex.org/W4321444361 |
| fwci | 0.0 |
| type | preprint |
| title | A Simple Way to Incorporate Target Structural Information in Molecular Generative Models |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T11948 |
| topics[1].field.id | https://openalex.org/fields/25 |
| topics[1].field.display_name | Materials Science |
| topics[1].score | 0.996399998664856 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2505 |
| topics[1].subfield.display_name | Materials Chemistry |
| topics[1].display_name | Machine Learning in Materials Science |
| topics[2].id | https://openalex.org/T10044 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9904000163078308 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Protein Structure and Dynamics |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C41008148 |
| concepts[0].level | 0 |
| concepts[0].score | 0.7176330089569092 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[0].display_name | Computer science |
| concepts[1].id | https://openalex.org/C39890363 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6856445074081421 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q36108 |
| concepts[1].display_name | Generative grammar |
| concepts[2].id | https://openalex.org/C154945302 |
| concepts[2].level | 1 |
| concepts[2].score | 0.6054113507270813 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[2].display_name | Artificial intelligence |
| concepts[3].id | https://openalex.org/C50644808 |
| concepts[3].level | 2 |
| concepts[3].score | 0.595265805721283 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q192776 |
| concepts[3].display_name | Artificial neural network |
| concepts[4].id | https://openalex.org/C167966045 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5653889775276184 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q5532625 |
| concepts[4].display_name | Generative model |
| concepts[5].id | https://openalex.org/C119857082 |
| concepts[5].level | 1 |
| concepts[5].score | 0.5612283945083618 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[5].display_name | Machine learning |
| concepts[6].id | https://openalex.org/C99726746 |
| concepts[6].level | 3 |
| concepts[6].score | 0.5413150787353516 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q906396 |
| concepts[6].display_name | Chemical space |
| concepts[7].id | https://openalex.org/C41685203 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4800930321216583 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1974042 |
| concepts[7].display_name | Docking (animal) |
| concepts[8].id | https://openalex.org/C61272859 |
| concepts[8].level | 3 |
| concepts[8].score | 0.4604000151157379 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7834031 |
| concepts[8].display_name | Transferability |
| concepts[9].id | https://openalex.org/C51632099 |
| concepts[9].level | 2 |
| concepts[9].score | 0.4602597653865814 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q3985153 |
| concepts[9].display_name | Training set |
| concepts[10].id | https://openalex.org/C74187038 |
| concepts[10].level | 2 |
| concepts[10].score | 0.44364726543426514 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q1418791 |
| concepts[10].display_name | Drug discovery |
| concepts[11].id | https://openalex.org/C2989108626 |
| concepts[11].level | 2 |
| concepts[11].score | 0.43920397758483887 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q904407 |
| concepts[11].display_name | Drug target |
| concepts[12].id | https://openalex.org/C185592680 |
| concepts[12].level | 0 |
| concepts[12].score | 0.1650504171848297 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[12].display_name | Chemistry |
| concepts[13].id | https://openalex.org/C60644358 |
| concepts[13].level | 1 |
| concepts[13].score | 0.15892231464385986 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[13].display_name | Bioinformatics |
| concepts[14].id | https://openalex.org/C86803240 |
| concepts[14].level | 0 |
| concepts[14].score | 0.08437585830688477 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[14].display_name | Biology |
| concepts[15].id | https://openalex.org/C55493867 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[15].display_name | Biochemistry |
| concepts[16].id | https://openalex.org/C71924100 |
| concepts[16].level | 0 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[16].display_name | Medicine |
| concepts[17].id | https://openalex.org/C159110408 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q121176 |
| concepts[17].display_name | Nursing |
| concepts[18].id | https://openalex.org/C140331021 |
| concepts[18].level | 2 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q1868104 |
| concepts[18].display_name | Logit |
| keywords[0].id | https://openalex.org/keywords/computer-science |
| keywords[0].score | 0.7176330089569092 |
| keywords[0].display_name | Computer science |
| keywords[1].id | https://openalex.org/keywords/generative-grammar |
| keywords[1].score | 0.6856445074081421 |
| keywords[1].display_name | Generative grammar |
| keywords[2].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[2].score | 0.6054113507270813 |
| keywords[2].display_name | Artificial intelligence |
| keywords[3].id | https://openalex.org/keywords/artificial-neural-network |
| keywords[3].score | 0.595265805721283 |
| keywords[3].display_name | Artificial neural network |
| keywords[4].id | https://openalex.org/keywords/generative-model |
| keywords[4].score | 0.5653889775276184 |
| keywords[4].display_name | Generative model |
| keywords[5].id | https://openalex.org/keywords/machine-learning |
| keywords[5].score | 0.5612283945083618 |
| keywords[5].display_name | Machine learning |
| keywords[6].id | https://openalex.org/keywords/chemical-space |
| keywords[6].score | 0.5413150787353516 |
| keywords[6].display_name | Chemical space |
| keywords[7].id | https://openalex.org/keywords/docking |
| keywords[7].score | 0.4800930321216583 |
| keywords[7].display_name | Docking (animal) |
| keywords[8].id | https://openalex.org/keywords/transferability |
| keywords[8].score | 0.4604000151157379 |
| keywords[8].display_name | Transferability |
| keywords[9].id | https://openalex.org/keywords/training-set |
| keywords[9].score | 0.4602597653865814 |
| keywords[9].display_name | Training set |
| keywords[10].id | https://openalex.org/keywords/drug-discovery |
| keywords[10].score | 0.44364726543426514 |
| keywords[10].display_name | Drug discovery |
| keywords[11].id | https://openalex.org/keywords/drug-target |
| keywords[11].score | 0.43920397758483887 |
| keywords[11].display_name | Drug target |
| keywords[12].id | https://openalex.org/keywords/chemistry |
| keywords[12].score | 0.1650504171848297 |
| keywords[12].display_name | Chemistry |
| keywords[13].id | https://openalex.org/keywords/bioinformatics |
| keywords[13].score | 0.15892231464385986 |
| keywords[13].display_name | Bioinformatics |
| keywords[14].id | https://openalex.org/keywords/biology |
| keywords[14].score | 0.08437585830688477 |
| keywords[14].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1101/2023.02.17.529000 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.02.17.529000 |
| locations[1].id | pmh:oai:figshare.com:article/23524254 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400572 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[1].source.host_organization | https://openalex.org/I196829312 |
| locations[1].source.host_organization_name | La Trobe University |
| locations[1].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[1].license | cc-by-nc |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Text |
| locations[1].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://figshare.com/articles/journal_contribution/A_Simple_Way_to_Incorporate_Target_Structural_Information_in_Molecular_Generative_Models/23524254 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5100360025 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-5514-9676 |
| authorships[0].author.display_name | Wenyi Zhang |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I3133055985 |
| authorships[0].affiliations[0].raw_affiliation_string | Institute of Biology, Westlake Institute for Advanced Study, 18 Shilongshan Road, Hangzhou, Zhejiang 10024, China |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I3133055985 |
| authorships[0].affiliations[1].raw_affiliation_string | Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang 31004, China |
| authorships[0].affiliations[2].institution_ids | https://openalex.org/I3133055985 |
| authorships[0].affiliations[2].raw_affiliation_string | Westlake AI Therapeutics Lab, Westlake Laboratory of Life Sciences and Biomedicine, 8 Shilongshan Road, Hangzhou, Zhejiang 30024, China |
| authorships[0].institutions[0].id | https://openalex.org/I3133055985 |
| authorships[0].institutions[0].ror | https://ror.org/05hfa4n20 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I3133055985 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Westlake University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Wenyi Zhang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Institute of Biology, Westlake Institute for Advanced Study, 18 Shilongshan Road, Hangzhou, Zhejiang 10024, China, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang 31004, China, Westlake AI Therapeutics Lab, Westlake Laboratory of Life Sciences and Biomedicine, 8 Shilongshan Road, Hangzhou, Zhejiang 30024, China |
| authorships[1].author.id | https://openalex.org/A5025284426 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3291-2015 |
| authorships[1].author.display_name | Kaiyue Zhang |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I3133055985 |
| authorships[1].affiliations[0].raw_affiliation_string | Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang 31004, China |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I3133055985 |
| authorships[1].affiliations[1].raw_affiliation_string | Westlake AI Therapeutics Lab, Westlake Laboratory of Life Sciences and Biomedicine, 8 Shilongshan Road, Hangzhou, Zhejiang 30024, China |
| authorships[1].institutions[0].id | https://openalex.org/I3133055985 |
| authorships[1].institutions[0].ror | https://ror.org/05hfa4n20 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I3133055985 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Westlake University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Kaiyue Zhang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang 31004, China, Westlake AI Therapeutics Lab, Westlake Laboratory of Life Sciences and Biomedicine, 8 Shilongshan Road, Hangzhou, Zhejiang 30024, China |
| authorships[2].author.id | https://openalex.org/A5060822545 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-9639-2907 |
| authorships[2].author.display_name | Jing Huang |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I3133055985 |
| authorships[2].affiliations[0].raw_affiliation_string | Institute of Biology, Westlake Institute for Advanced Study, 18 Shilongshan Road, Hangzhou, Zhejiang 10024, China |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I3133055985 |
| authorships[2].affiliations[1].raw_affiliation_string | Westlake AI Therapeutics Lab, Westlake Laboratory of Life Sciences and Biomedicine, 8 Shilongshan Road, Hangzhou, Zhejiang 30024, China |
| authorships[2].affiliations[2].institution_ids | https://openalex.org/I3133055985 |
| authorships[2].affiliations[2].raw_affiliation_string | Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang 31004, China |
| authorships[2].institutions[0].id | https://openalex.org/I3133055985 |
| authorships[2].institutions[0].ror | https://ror.org/05hfa4n20 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I3133055985 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Westlake University |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Jing Huang |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Institute of Biology, Westlake Institute for Advanced Study, 18 Shilongshan Road, Hangzhou, Zhejiang 10024, China, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang 31004, China, Westlake AI Therapeutics Lab, Westlake Laboratory of Life Sciences and Biomedicine, 8 Shilongshan Road, Hangzhou, Zhejiang 30024, China |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | A Simple Way to Incorporate Target Structural Information in Molecular Generative Models |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W4237416477, https://openalex.org/W3127641366, https://openalex.org/W3185391990, https://openalex.org/W2952410321, https://openalex.org/W2765324078, https://openalex.org/W2798505693, https://openalex.org/W4388265932, https://openalex.org/W4306801745, https://openalex.org/W2219197936, https://openalex.org/W2059632225 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1101/2023.02.17.529000 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.02.17.529000 |
| primary_location.id | doi:10.1101/2023.02.17.529000 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/05/31/2023.02.17.529000.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.02.17.529000 |
| publication_date | 2023-02-21 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W3201539868, https://openalex.org/W3020511336, https://openalex.org/W2912171584, https://openalex.org/W4210589602, https://openalex.org/W4200233821, https://openalex.org/W4210617107, https://openalex.org/W4297995910, https://openalex.org/W2017254234, https://openalex.org/W2151971404, https://openalex.org/W4304203195, https://openalex.org/W3209056694, https://openalex.org/W2991736596, https://openalex.org/W2989615256, https://openalex.org/W2773987374, https://openalex.org/W2794063970, https://openalex.org/W4309413039, https://openalex.org/W3133989558, https://openalex.org/W3162583919, https://openalex.org/W3109549311, https://openalex.org/W3125566655, https://openalex.org/W3200806939, https://openalex.org/W3209764902, https://openalex.org/W4226383578, https://openalex.org/W3213129754, https://openalex.org/W3135642867, https://openalex.org/W3112474878, https://openalex.org/W3144567229, https://openalex.org/W3027975282, https://openalex.org/W4200150297, https://openalex.org/W2134967712, https://openalex.org/W2299115575, https://openalex.org/W3094686696, https://openalex.org/W2610148085, https://openalex.org/W1966613725, https://openalex.org/W2066273100, https://openalex.org/W3090915092, https://openalex.org/W2900090807, https://openalex.org/W3094640617, https://openalex.org/W2789748145, https://openalex.org/W1997176810, https://openalex.org/W3209118444, https://openalex.org/W3106761016, https://openalex.org/W2334483166, https://openalex.org/W2128332459, https://openalex.org/W2388080941, https://openalex.org/W3092297806, https://openalex.org/W4296794490, https://openalex.org/W3029978182, https://openalex.org/W3174777205, https://openalex.org/W4221054065, https://openalex.org/W2966357564, https://openalex.org/W2108389384, https://openalex.org/W2067643341, https://openalex.org/W2105668062, https://openalex.org/W2042572511, https://openalex.org/W3159598019, https://openalex.org/W3100751385 |
| referenced_works_count | 57 |
| abstract_inverted_index.A | 74 |
| abstract_inverted_index.a | 20, 40, 50, 71, 100, 137, 171 |
| abstract_inverted_index.3D | 26 |
| abstract_inverted_index.In | 15 |
| abstract_inverted_index.as | 55 |
| abstract_inverted_index.in | 9, 29, 140 |
| abstract_inverted_index.is | 80 |
| abstract_inverted_index.of | 77, 83, 95, 110, 119 |
| abstract_inverted_index.on | 116, 132 |
| abstract_inverted_index.or | 158 |
| abstract_inverted_index.to | 23, 59, 90, 144, 151, 155 |
| abstract_inverted_index.we | 18 |
| abstract_inverted_index.The | 37 |
| abstract_inverted_index.and | 124, 148, 173 |
| abstract_inverted_index.are | 5 |
| abstract_inverted_index.for | 33, 65, 87, 127, 162, 177 |
| abstract_inverted_index.hit | 141 |
| abstract_inverted_index.its | 56 |
| abstract_inverted_index.key | 75 |
| abstract_inverted_index.now | 6 |
| abstract_inverted_index.the | 61, 78, 81, 111, 117, 128, 149 |
| abstract_inverted_index.Deep | 1 |
| abstract_inverted_index.This | 168 |
| abstract_inverted_index.bind | 68 |
| abstract_inverted_index.drug | 13, 35 |
| abstract_inverted_index.sets | 86 |
| abstract_inverted_index.that | 45, 67 |
| abstract_inverted_index.this | 16, 105 |
| abstract_inverted_index.with | 49, 70 |
| abstract_inverted_index.Tests | 131 |
| abstract_inverted_index.about | 122 |
| abstract_inverted_index.being | 7 |
| abstract_inverted_index.drugs | 157 |
| abstract_inverted_index.eight | 133 |
| abstract_inverted_index.known | 159 |
| abstract_inverted_index.model | 44, 54 |
| abstract_inverted_index.novel | 21 |
| abstract_inverted_index.prior | 120, 166 |
| abstract_inverted_index.space | 63, 113 |
| abstract_inverted_index.work, | 17 |
| abstract_inverted_index.active | 123, 160 |
| abstract_inverted_index.fields | 11 |
| abstract_inverted_index.guided | 108 |
| abstract_inverted_index.highly | 174 |
| abstract_inverted_index.issues | 94 |
| abstract_inverted_index.method | 38, 79, 169 |
| abstract_inverted_index.models | 4, 32, 98 |
| abstract_inverted_index.neural | 42, 52 |
| abstract_inverted_index.reward | 57 |
| abstract_inverted_index.scores | 48 |
| abstract_inverted_index.showed | 136 |
| abstract_inverted_index.target | 25, 134 |
| abstract_inverted_index.ability | 150 |
| abstract_inverted_index.applied | 8 |
| abstract_inverted_index.design. | 36 |
| abstract_inverted_index.docking | 47, 97, 146 |
| abstract_inverted_index.enables | 106 |
| abstract_inverted_index.feature | 76 |
| abstract_inverted_index.general | 172 |
| abstract_inverted_index.include | 24 |
| abstract_inverted_index.ligands | 161 |
| abstract_inverted_index.network | 43, 53 |
| abstract_inverted_index.propose | 19 |
| abstract_inverted_index.similar | 154 |
| abstract_inverted_index.target. | 73, 130 |
| abstract_inverted_index.targets | 164 |
| abstract_inverted_index.through | 99 |
| abstract_inverted_index.various | 10 |
| abstract_inverted_index.without | 114, 165 |
| abstract_inverted_index.100-fold | 138 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.accurate | 107 |
| abstract_inverted_index.approach | 22 |
| abstract_inverted_index.approved | 156 |
| abstract_inverted_index.chemical | 62, 112 |
| abstract_inverted_index.combines | 39 |
| abstract_inverted_index.compared | 143 |
| abstract_inverted_index.designed | 89 |
| abstract_inverted_index.function | 58 |
| abstract_inverted_index.generate | 152 |
| abstract_inverted_index.inactive | 125 |
| abstract_inverted_index.increase | 139 |
| abstract_inverted_index.learning | 2 |
| abstract_inverted_index.navigate | 60 |
| abstract_inverted_index.overcome | 91 |
| abstract_inverted_index.predicts | 46 |
| abstract_inverted_index.process. | 103 |
| abstract_inverted_index.proteins | 135 |
| abstract_inverted_index.provides | 170 |
| abstract_inverted_index.reliance | 115 |
| abstract_inverted_index.solution | 176 |
| abstract_inverted_index.specific | 72, 129, 163 |
| abstract_inverted_index.training | 102 |
| abstract_inverted_index.compounds | 126 |
| abstract_inverted_index.efficient | 175 |
| abstract_inverted_index.favorably | 69 |
| abstract_inverted_index.including | 12 |
| abstract_inverted_index.knowledge | 121 |
| abstract_inverted_index.molecular | 30, 85, 179 |
| abstract_inverted_index.molecules | 66, 153 |
| abstract_inverted_index.potential | 92 |
| abstract_inverted_index.searching | 64 |
| abstract_inverted_index.surrogate | 96 |
| abstract_inverted_index.training, | 88 |
| abstract_inverted_index.two-round | 101 |
| abstract_inverted_index.collection | 118 |
| abstract_inverted_index.discovery. | 14 |
| abstract_inverted_index.generation | 142 |
| abstract_inverted_index.generative | 3, 31, 51 |
| abstract_inverted_index.knowledge. | 167 |
| abstract_inverted_index.structural | 27 |
| abstract_inverted_index.exploration | 109 |
| abstract_inverted_index.generation. | 180 |
| abstract_inverted_index.information | 28 |
| abstract_inverted_index.construction | 82 |
| abstract_inverted_index.conventional | 145 |
| abstract_inverted_index.Consequently, | 104 |
| abstract_inverted_index.calculations, | 147 |
| abstract_inverted_index.message-passing | 41 |
| abstract_inverted_index.structure-based | 34, 178 |
| abstract_inverted_index.target-specific | 84 |
| abstract_inverted_index.transferability | 93 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5060822545 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I3133055985 |
| citation_normalized_percentile.value | 0.03449359 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |