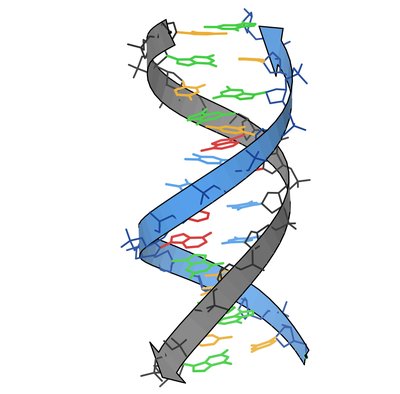
A Tale of Loops and Tails: The Role of Intrinsically Disordered Protein Regions in R-Loop Recognition and Phase Separation Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3389/fmolb.2021.691694
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3389/fmolb.2021.691694
R-loops are non-canonical, three-stranded nucleic acid structures composed of a DNA:RNA hybrid, a displaced single-stranded (ss)DNA, and a trailing ssRNA overhang. R-loops perform critical biological functions under both normal and disease conditions. To elucidate their cellular functions, we need to understand the mechanisms underlying R-loop formation, recognition, signaling, and resolution. Previous high-throughput screens identified multiple proteins that bind R-loops, with many of these proteins containing folded nucleic acid processing and binding domains that prevent (e.g., topoisomerases), resolve (e.g., helicases, nucleases), or recognize (e.g., KH, RRMs) R-loops. However, a significant number of these R-loop interacting Enzyme and Reader proteins also contain long stretches of intrinsically disordered regions (IDRs). The precise molecular and structural mechanisms by which the folded domains and IDRs synergize to recognize and process R-loops or modulate R-loop-mediated signaling have not been fully explored. While studying one such modular R-loop Reader, the Fragile X Protein (FMRP), we unexpectedly discovered that the C-terminal IDR (C-IDR) of FMRP is the predominant R-loop binding site, with the three N-terminal KH domains recognizing the trailing ssRNA overhang. Interestingly, the C-IDR of FMRP has recently been shown to undergo spontaneous Liquid-Liquid Phase Separation (LLPS) assembly by itself or in complex with another non-canonical nucleic acid structure, RNA G-quadruplex. Furthermore, we have recently shown that FMRP can suppress persistent R-loops that form during transcription, a process that is also enhanced by LLPS via the assembly of membraneless transcription factories. These exciting findings prompted us to explore the role of IDRs in R-loop processing and signaling proteins through a comprehensive bioinformatics and computational biology study. Here, we evaluated IDR prevalence, sequence composition and LLPS propensity for the known R-loop interactome. We observed that, like FMRP, the majority of the R-loop interactome, especially Readers, contains long IDRs that are highly enriched in low complexity sequences with biased amino acid composition, suggesting that these IDRs could directly interact with R-loops, rather than being “mere flexible linkers” connecting the “functional folded enzyme or binding domains”. Furthermore, our analysis shows that several proteins in the R-loop interactome are either predicted to or have been experimentally demonstrated to undergo LLPS or are known to be associated with phase separated membraneless organelles. Thus, our overall results present a thought-provoking hypothesis that IDRs in the R-loop interactome can provide a functional link between R-loop recognition via direct binding and downstream signaling through the assembly of LLPS-mediated membrane-less R-loop foci. The absence or dysregulation of the function of IDR-enriched R-loop interactors can potentially lead to severe genomic defects, such as the widespread R-loop-mediated DNA double strand breaks that we recently observed in Fragile X patient-derived cells.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3389/fmolb.2021.691694
- OA Status
- gold
- Cited By
- 34
- References
- 136
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3169357026
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3169357026Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3389/fmolb.2021.691694Digital Object Identifier
- Title
-
A Tale of Loops and Tails: The Role of Intrinsically Disordered Protein Regions in R-Loop Recognition and Phase SeparationWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-06-10Full publication date if available
- Authors
-
Leonardo G. Dettori, Diego Torrejon, Arijita Chakraborty, Arijit Dutta, Mohamed Mohamed, Csaba Papp, Vladimir A. Kuznetsov, Patrick Sung, Wenyi Feng, Alaji BahList of authors in order
- Landing page
-
https://doi.org/10.3389/fmolb.2021.691694Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.3389/fmolb.2021.691694Direct OA link when available
- Concepts
-
Nucleic acid, RNA, DNA, Loop (graph theory), Biology, Biophysics, Helicase, Intrinsically disordered proteins, Chemistry, Cell biology, Biochemistry, Gene, Mathematics, CombinatoricsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
34Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 7, 2024: 6, 2023: 6, 2022: 11, 2021: 4Per-year citation counts (last 5 years)
- References (count)
-
136Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3169357026 |
|---|---|
| doi | https://doi.org/10.3389/fmolb.2021.691694 |
| ids.doi | https://doi.org/10.3389/fmolb.2021.691694 |
| ids.mag | 3169357026 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/34179096 |
| ids.openalex | https://openalex.org/W3169357026 |
| fwci | 2.63337554 |
| type | article |
| title | A Tale of Loops and Tails: The Role of Intrinsically Disordered Protein Regions in R-Loop Recognition and Phase Separation |
| biblio.issue | |
| biblio.volume | 8 |
| biblio.last_page | 691694 |
| biblio.first_page | 691694 |
| topics[0].id | https://openalex.org/T10604 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9995999932289124 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | RNA Research and Splicing |
| topics[1].id | https://openalex.org/T11482 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9994999766349792 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | RNA modifications and cancer |
| topics[2].id | https://openalex.org/T10521 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9991999864578247 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA and protein synthesis mechanisms |
| is_xpac | False |
| apc_list.value | 2490 |
| apc_list.currency | USD |
| apc_list.value_usd | 2490 |
| apc_paid.value | 2490 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2490 |
| concepts[0].id | https://openalex.org/C24107716 |
| concepts[0].level | 2 |
| concepts[0].score | 0.5796681046485901 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q123619 |
| concepts[0].display_name | Nucleic acid |
| concepts[1].id | https://openalex.org/C67705224 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5627881288528442 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q11053 |
| concepts[1].display_name | RNA |
| concepts[2].id | https://openalex.org/C552990157 |
| concepts[2].level | 2 |
| concepts[2].score | 0.4990353584289551 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[2].display_name | DNA |
| concepts[3].id | https://openalex.org/C184670325 |
| concepts[3].level | 2 |
| concepts[3].score | 0.4937179982662201 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q512604 |
| concepts[3].display_name | Loop (graph theory) |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.48335498571395874 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C12554922 |
| concepts[5].level | 1 |
| concepts[5].score | 0.46776291728019714 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7100 |
| concepts[5].display_name | Biophysics |
| concepts[6].id | https://openalex.org/C161223559 |
| concepts[6].level | 4 |
| concepts[6].score | 0.43349146842956543 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q138864 |
| concepts[6].display_name | Helicase |
| concepts[7].id | https://openalex.org/C2778815515 |
| concepts[7].level | 2 |
| concepts[7].score | 0.42603519558906555 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q3408242 |
| concepts[7].display_name | Intrinsically disordered proteins |
| concepts[8].id | https://openalex.org/C185592680 |
| concepts[8].level | 0 |
| concepts[8].score | 0.3622720241546631 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[8].display_name | Chemistry |
| concepts[9].id | https://openalex.org/C95444343 |
| concepts[9].level | 1 |
| concepts[9].score | 0.32055580615997314 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[9].display_name | Cell biology |
| concepts[10].id | https://openalex.org/C55493867 |
| concepts[10].level | 1 |
| concepts[10].score | 0.29391443729400635 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[10].display_name | Biochemistry |
| concepts[11].id | https://openalex.org/C104317684 |
| concepts[11].level | 2 |
| concepts[11].score | 0.08720704913139343 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[11].display_name | Gene |
| concepts[12].id | https://openalex.org/C33923547 |
| concepts[12].level | 0 |
| concepts[12].score | 0.0 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[12].display_name | Mathematics |
| concepts[13].id | https://openalex.org/C114614502 |
| concepts[13].level | 1 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q76592 |
| concepts[13].display_name | Combinatorics |
| keywords[0].id | https://openalex.org/keywords/nucleic-acid |
| keywords[0].score | 0.5796681046485901 |
| keywords[0].display_name | Nucleic acid |
| keywords[1].id | https://openalex.org/keywords/rna |
| keywords[1].score | 0.5627881288528442 |
| keywords[1].display_name | RNA |
| keywords[2].id | https://openalex.org/keywords/dna |
| keywords[2].score | 0.4990353584289551 |
| keywords[2].display_name | DNA |
| keywords[3].id | https://openalex.org/keywords/loop |
| keywords[3].score | 0.4937179982662201 |
| keywords[3].display_name | Loop (graph theory) |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.48335498571395874 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/biophysics |
| keywords[5].score | 0.46776291728019714 |
| keywords[5].display_name | Biophysics |
| keywords[6].id | https://openalex.org/keywords/helicase |
| keywords[6].score | 0.43349146842956543 |
| keywords[6].display_name | Helicase |
| keywords[7].id | https://openalex.org/keywords/intrinsically-disordered-proteins |
| keywords[7].score | 0.42603519558906555 |
| keywords[7].display_name | Intrinsically disordered proteins |
| keywords[8].id | https://openalex.org/keywords/chemistry |
| keywords[8].score | 0.3622720241546631 |
| keywords[8].display_name | Chemistry |
| keywords[9].id | https://openalex.org/keywords/cell-biology |
| keywords[9].score | 0.32055580615997314 |
| keywords[9].display_name | Cell biology |
| keywords[10].id | https://openalex.org/keywords/biochemistry |
| keywords[10].score | 0.29391443729400635 |
| keywords[10].display_name | Biochemistry |
| keywords[11].id | https://openalex.org/keywords/gene |
| keywords[11].score | 0.08720704913139343 |
| keywords[11].display_name | Gene |
| language | en |
| locations[0].id | doi:10.3389/fmolb.2021.691694 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2595797843 |
| locations[0].source.issn | 2296-889X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2296-889X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Frontiers in Molecular Biosciences |
| locations[0].source.host_organization | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_name | Frontiers Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_lineage_names | Frontiers Media |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Frontiers in Molecular Biosciences |
| locations[0].landing_page_url | https://doi.org/10.3389/fmolb.2021.691694 |
| locations[1].id | pmid:34179096 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Frontiers in molecular biosciences |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/34179096 |
| locations[2].id | pmh:oai:doaj.org/article:3bb66076549b4dee8876f201be381aaa |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Frontiers in Molecular Biosciences, Vol 8 (2021) |
| locations[2].landing_page_url | https://doaj.org/article/3bb66076549b4dee8876f201be381aaa |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:8222781 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Front Mol Biosci |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/8222781 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5058714251 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-7333-3584 |
| authorships[0].author.display_name | Leonardo G. Dettori |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[0].institutions[0].id | https://openalex.org/I20388574 |
| authorships[0].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Leonardo G. Dettori |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[1].author.id | https://openalex.org/A5085596190 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Diego Torrejon |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[1].institutions[0].id | https://openalex.org/I20388574 |
| authorships[1].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Diego Torrejon |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[2].author.id | https://openalex.org/A5000419914 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-4616-4399 |
| authorships[2].author.display_name | Arijita Chakraborty |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[2].institutions[0].id | https://openalex.org/I20388574 |
| authorships[2].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Arijita Chakraborty |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[3].author.id | https://openalex.org/A5071589025 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-5675-0092 |
| authorships[3].author.display_name | Arijit Dutta |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I165951966, https://openalex.org/I45438204 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Biochemistry and Structural Biology, University of Texas Health San Antonio, San Antonio, TX, United States |
| authorships[3].institutions[0].id | https://openalex.org/I165951966 |
| authorships[3].institutions[0].ror | https://ror.org/02f6dcw23 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I165951966 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | The University of Texas Health Science Center at San Antonio |
| authorships[3].institutions[1].id | https://openalex.org/I45438204 |
| authorships[3].institutions[1].ror | https://ror.org/01kd65564 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I45438204 |
| authorships[3].institutions[1].country_code | US |
| authorships[3].institutions[1].display_name | The University of Texas at San Antonio |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Arijit Dutta |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Biochemistry and Structural Biology, University of Texas Health San Antonio, San Antonio, TX, United States |
| authorships[4].author.id | https://openalex.org/A5104560224 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Mohamed Mohamed |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[4].institutions[0].id | https://openalex.org/I20388574 |
| authorships[4].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Mohamed Mohamed |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[5].author.id | https://openalex.org/A5042922889 |
| authorships[5].author.orcid | https://orcid.org/0000-0003-3874-0162 |
| authorships[5].author.display_name | Csaba Papp |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Urology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I20388574 |
| authorships[5].affiliations[1].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[5].institutions[0].id | https://openalex.org/I20388574 |
| authorships[5].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Csaba Papp |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States, Department of Urology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[6].author.id | https://openalex.org/A5101607058 |
| authorships[6].author.orcid | https://orcid.org/0000-0003-3405-0762 |
| authorships[6].author.display_name | Vladimir A. Kuznetsov |
| authorships[6].countries | SG, US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Urology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I115228651, https://openalex.org/I4210148498 |
| authorships[6].affiliations[1].raw_affiliation_string | Bioinformatics Institute, A*STAR Biomedical Institutes, Singapore, Singapore |
| authorships[6].institutions[0].id | https://openalex.org/I115228651 |
| authorships[6].institutions[0].ror | https://ror.org/036wvzt09 |
| authorships[6].institutions[0].type | government |
| authorships[6].institutions[0].lineage | https://openalex.org/I115228651 |
| authorships[6].institutions[0].country_code | SG |
| authorships[6].institutions[0].display_name | Agency for Science, Technology and Research |
| authorships[6].institutions[1].id | https://openalex.org/I4210148498 |
| authorships[6].institutions[1].ror | https://ror.org/044w3nw43 |
| authorships[6].institutions[1].type | facility |
| authorships[6].institutions[1].lineage | https://openalex.org/I115228651, https://openalex.org/I2801752549, https://openalex.org/I4210148498 |
| authorships[6].institutions[1].country_code | SG |
| authorships[6].institutions[1].display_name | Bioinformatics Institute |
| authorships[6].institutions[2].id | https://openalex.org/I20388574 |
| authorships[6].institutions[2].ror | https://ror.org/040kfrw16 |
| authorships[6].institutions[2].type | education |
| authorships[6].institutions[2].lineage | https://openalex.org/I20388574 |
| authorships[6].institutions[2].country_code | US |
| authorships[6].institutions[2].display_name | SUNY Upstate Medical University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Vladimir A. Kuznetsov |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Bioinformatics Institute, A*STAR Biomedical Institutes, Singapore, Singapore, Department of Urology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[7].author.id | https://openalex.org/A5090899284 |
| authorships[7].author.orcid | https://orcid.org/0000-0003-1396-9040 |
| authorships[7].author.display_name | Patrick Sung |
| authorships[7].countries | US |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I165951966, https://openalex.org/I45438204 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Biochemistry and Structural Biology, University of Texas Health San Antonio, San Antonio, TX, United States |
| authorships[7].institutions[0].id | https://openalex.org/I165951966 |
| authorships[7].institutions[0].ror | https://ror.org/02f6dcw23 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I165951966 |
| authorships[7].institutions[0].country_code | US |
| authorships[7].institutions[0].display_name | The University of Texas Health Science Center at San Antonio |
| authorships[7].institutions[1].id | https://openalex.org/I45438204 |
| authorships[7].institutions[1].ror | https://ror.org/01kd65564 |
| authorships[7].institutions[1].type | education |
| authorships[7].institutions[1].lineage | https://openalex.org/I45438204 |
| authorships[7].institutions[1].country_code | US |
| authorships[7].institutions[1].display_name | The University of Texas at San Antonio |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Patrick Sung |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Department of Biochemistry and Structural Biology, University of Texas Health San Antonio, San Antonio, TX, United States |
| authorships[8].author.id | https://openalex.org/A5012168976 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-6123-7998 |
| authorships[8].author.display_name | Wenyi Feng |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[8].institutions[0].id | https://openalex.org/I20388574 |
| authorships[8].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Wenyi Feng |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[9].author.id | https://openalex.org/A5086367121 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-8178-0402 |
| authorships[9].author.display_name | Alaji Bah |
| authorships[9].countries | US |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I20388574 |
| authorships[9].affiliations[0].raw_affiliation_string | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| authorships[9].institutions[0].id | https://openalex.org/I20388574 |
| authorships[9].institutions[0].ror | https://ror.org/040kfrw16 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I20388574 |
| authorships[9].institutions[0].country_code | US |
| authorships[9].institutions[0].display_name | SUNY Upstate Medical University |
| authorships[9].author_position | last |
| authorships[9].raw_author_name | Alaji Bah |
| authorships[9].is_corresponding | True |
| authorships[9].raw_affiliation_strings | Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, United States |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.3389/fmolb.2021.691694 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | A Tale of Loops and Tails: The Role of Intrinsically Disordered Protein Regions in R-Loop Recognition and Phase Separation |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10604 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9995999932289124 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | RNA Research and Splicing |
| related_works | https://openalex.org/W4293203577, https://openalex.org/W3198650065, https://openalex.org/W2160213949, https://openalex.org/W4288038979, https://openalex.org/W2516052007, https://openalex.org/W1971444612, https://openalex.org/W2964159616, https://openalex.org/W1543504406, https://openalex.org/W2161208925, https://openalex.org/W2969721268 |
| cited_by_count | 34 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 7 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 6 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 6 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 11 |
| counts_by_year[4].year | 2021 |
| counts_by_year[4].cited_by_count | 4 |
| locations_count | 4 |
| best_oa_location.id | doi:10.3389/fmolb.2021.691694 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2595797843 |
| best_oa_location.source.issn | 2296-889X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2296-889X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Frontiers in Molecular Biosciences |
| best_oa_location.source.host_organization | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_name | Frontiers Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_lineage_names | Frontiers Media |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Frontiers in Molecular Biosciences |
| best_oa_location.landing_page_url | https://doi.org/10.3389/fmolb.2021.691694 |
| primary_location.id | doi:10.3389/fmolb.2021.691694 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2595797843 |
| primary_location.source.issn | 2296-889X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2296-889X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Frontiers in Molecular Biosciences |
| primary_location.source.host_organization | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_name | Frontiers Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_lineage_names | Frontiers Media |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Frontiers in Molecular Biosciences |
| primary_location.landing_page_url | https://doi.org/10.3389/fmolb.2021.691694 |
| publication_date | 2021-06-10 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W1975993845, https://openalex.org/W2091796431, https://openalex.org/W2110932617, https://openalex.org/W2126099124, https://openalex.org/W2919749323, https://openalex.org/W2906567604, https://openalex.org/W2036338927, https://openalex.org/W2085002173, https://openalex.org/W2085656056, https://openalex.org/W2042745266, https://openalex.org/W2042409563, https://openalex.org/W4247091049, https://openalex.org/W3125860677, https://openalex.org/W2077205550, https://openalex.org/W2150636365, https://openalex.org/W2901209536, https://openalex.org/W2425405879, https://openalex.org/W2087587381, https://openalex.org/W2955067398, https://openalex.org/W2010458140, https://openalex.org/W2027657574, https://openalex.org/W1970383168, https://openalex.org/W2098756736, https://openalex.org/W2131110198, https://openalex.org/W1557728494, https://openalex.org/W3153641076, https://openalex.org/W3088651435, https://openalex.org/W1964680558, https://openalex.org/W2097627856, https://openalex.org/W2798187866, https://openalex.org/W2885156893, https://openalex.org/W2800358055, https://openalex.org/W2972330631, https://openalex.org/W2913346540, https://openalex.org/W3036292274, https://openalex.org/W2111375641, https://openalex.org/W2907750653, https://openalex.org/W2006192061, https://openalex.org/W2084728529, https://openalex.org/W3014133106, https://openalex.org/W2099650039, https://openalex.org/W2104301158, https://openalex.org/W2004342279, https://openalex.org/W2979531443, https://openalex.org/W2125761685, https://openalex.org/W1967706132, https://openalex.org/W2044375974, https://openalex.org/W2590327834, https://openalex.org/W2995711559, https://openalex.org/W2166434010, https://openalex.org/W2782562502, https://openalex.org/W2902044057, https://openalex.org/W3018445299, https://openalex.org/W2617594926, https://openalex.org/W2558179461, https://openalex.org/W2098090834, https://openalex.org/W2770128744, https://openalex.org/W2031767160, https://openalex.org/W4242223940, https://openalex.org/W2102027852, https://openalex.org/W2811123412, https://openalex.org/W2085978470, https://openalex.org/W2971594646, https://openalex.org/W1972011626, https://openalex.org/W2909481675, https://openalex.org/W2787093671, https://openalex.org/W3008165273, https://openalex.org/W2281827817, https://openalex.org/W3126870155, https://openalex.org/W2952073075, https://openalex.org/W2980646921, https://openalex.org/W2503054853, https://openalex.org/W2165608602, https://openalex.org/W2097892623, https://openalex.org/W2148816697, https://openalex.org/W2991067665, https://openalex.org/W2539844013, https://openalex.org/W2883403291, https://openalex.org/W2120683379, https://openalex.org/W1968593031, https://openalex.org/W2120745370, https://openalex.org/W2802015040, https://openalex.org/W2809057270, https://openalex.org/W3043233893, https://openalex.org/W2765869464, https://openalex.org/W1921890141, https://openalex.org/W2123824748, https://openalex.org/W2340421620, https://openalex.org/W2463453322, https://openalex.org/W1989540449, https://openalex.org/W2154968223, https://openalex.org/W2161887614, https://openalex.org/W2928730330, https://openalex.org/W1686075321, https://openalex.org/W2107455041, https://openalex.org/W2990332438, https://openalex.org/W2064990267, https://openalex.org/W2168272116, https://openalex.org/W3112376646, https://openalex.org/W2914590607, https://openalex.org/W2065884484, https://openalex.org/W2147733973, https://openalex.org/W2553996574, https://openalex.org/W2118608964, https://openalex.org/W1790543114, https://openalex.org/W1967566171, https://openalex.org/W1994840640, https://openalex.org/W2100232590, https://openalex.org/W2786469974, https://openalex.org/W2003881377, https://openalex.org/W2078403259, https://openalex.org/W2886727482, https://openalex.org/W2164332468, https://openalex.org/W2094384279, https://openalex.org/W2514004327, https://openalex.org/W3022023372, https://openalex.org/W1998706983, https://openalex.org/W2099009604, https://openalex.org/W183349502, https://openalex.org/W2803560469, https://openalex.org/W2080840166, https://openalex.org/W2739634185, https://openalex.org/W1970966311, https://openalex.org/W2945712825, https://openalex.org/W2772865572, https://openalex.org/W2987435148, https://openalex.org/W285093818, https://openalex.org/W2578577905, https://openalex.org/W2983578800, https://openalex.org/W2520321244, https://openalex.org/W2939780179, https://openalex.org/W2948704538, https://openalex.org/W2903630407, https://openalex.org/W2121942815, https://openalex.org/W2518509726, https://openalex.org/W3134294420 |
| referenced_works_count | 136 |
| abstract_inverted_index.X | 144, 429 |
| abstract_inverted_index.a | 9, 12, 17, 87, 219, 252, 365, 376 |
| abstract_inverted_index.KH | 167 |
| abstract_inverted_index.To | 32 |
| abstract_inverted_index.We | 274 |
| abstract_inverted_index.as | 415 |
| abstract_inverted_index.be | 353 |
| abstract_inverted_index.by | 113, 191, 225 |
| abstract_inverted_index.in | 194, 245, 294, 333, 370, 427 |
| abstract_inverted_index.is | 157, 222 |
| abstract_inverted_index.of | 8, 61, 90, 102, 155, 177, 230, 243, 281, 391, 400, 403 |
| abstract_inverted_index.or | 80, 126, 193, 323, 341, 349, 398 |
| abstract_inverted_index.to | 39, 121, 183, 239, 340, 346, 352, 410 |
| abstract_inverted_index.us | 238 |
| abstract_inverted_index.we | 37, 147, 205, 260, 424 |
| abstract_inverted_index.DNA | 419 |
| abstract_inverted_index.IDR | 153, 262 |
| abstract_inverted_index.KH, | 83 |
| abstract_inverted_index.RNA | 202 |
| abstract_inverted_index.The | 107, 396 |
| abstract_inverted_index.and | 16, 29, 48, 69, 95, 110, 118, 123, 248, 255, 266, 385 |
| abstract_inverted_index.are | 1, 291, 337, 350 |
| abstract_inverted_index.can | 211, 374, 407 |
| abstract_inverted_index.for | 269 |
| abstract_inverted_index.has | 179 |
| abstract_inverted_index.low | 295 |
| abstract_inverted_index.not | 131 |
| abstract_inverted_index.one | 137 |
| abstract_inverted_index.our | 327, 361 |
| abstract_inverted_index.the | 41, 115, 142, 151, 158, 164, 170, 175, 228, 241, 270, 279, 282, 319, 334, 371, 389, 401, 416 |
| abstract_inverted_index.via | 227, 382 |
| abstract_inverted_index.FMRP | 156, 178, 210 |
| abstract_inverted_index.IDRs | 119, 244, 289, 306, 369 |
| abstract_inverted_index.LLPS | 226, 267, 348 |
| abstract_inverted_index.acid | 5, 67, 200, 301 |
| abstract_inverted_index.also | 98, 223 |
| abstract_inverted_index.been | 132, 181, 343 |
| abstract_inverted_index.bind | 57 |
| abstract_inverted_index.both | 27 |
| abstract_inverted_index.form | 216 |
| abstract_inverted_index.have | 130, 206, 342 |
| abstract_inverted_index.lead | 409 |
| abstract_inverted_index.like | 277 |
| abstract_inverted_index.link | 378 |
| abstract_inverted_index.long | 100, 288 |
| abstract_inverted_index.many | 60 |
| abstract_inverted_index.need | 38 |
| abstract_inverted_index.role | 242 |
| abstract_inverted_index.such | 138, 414 |
| abstract_inverted_index.than | 313 |
| abstract_inverted_index.that | 56, 72, 150, 209, 215, 221, 290, 304, 330, 368, 423 |
| abstract_inverted_index.with | 59, 163, 196, 298, 310, 355 |
| abstract_inverted_index.C-IDR | 176 |
| abstract_inverted_index.FMRP, | 278 |
| abstract_inverted_index.Here, | 259 |
| abstract_inverted_index.Phase | 187 |
| abstract_inverted_index.RRMs) | 84 |
| abstract_inverted_index.These | 234 |
| abstract_inverted_index.Thus, | 360 |
| abstract_inverted_index.While | 135 |
| abstract_inverted_index.amino | 300 |
| abstract_inverted_index.being | 314 |
| abstract_inverted_index.could | 307 |
| abstract_inverted_index.foci. | 395 |
| abstract_inverted_index.fully | 133 |
| abstract_inverted_index.known | 271, 351 |
| abstract_inverted_index.phase | 356 |
| abstract_inverted_index.shown | 182, 208 |
| abstract_inverted_index.shows | 329 |
| abstract_inverted_index.site, | 162 |
| abstract_inverted_index.ssRNA | 19, 172 |
| abstract_inverted_index.that, | 276 |
| abstract_inverted_index.their | 34 |
| abstract_inverted_index.these | 62, 91, 305 |
| abstract_inverted_index.three | 165 |
| abstract_inverted_index.under | 26 |
| abstract_inverted_index.which | 114 |
| abstract_inverted_index.(LLPS) | 189 |
| abstract_inverted_index.(e.g., | 74, 77, 82 |
| abstract_inverted_index.Enzyme | 94 |
| abstract_inverted_index.R-loop | 44, 92, 140, 160, 246, 272, 283, 335, 372, 380, 394, 405 |
| abstract_inverted_index.Reader | 96 |
| abstract_inverted_index.biased | 299 |
| abstract_inverted_index.breaks | 422 |
| abstract_inverted_index.cells. | 431 |
| abstract_inverted_index.direct | 383 |
| abstract_inverted_index.double | 420 |
| abstract_inverted_index.during | 217 |
| abstract_inverted_index.either | 338 |
| abstract_inverted_index.enzyme | 322 |
| abstract_inverted_index.folded | 65, 116, 321 |
| abstract_inverted_index.highly | 292 |
| abstract_inverted_index.itself | 192 |
| abstract_inverted_index.normal | 28 |
| abstract_inverted_index.number | 89 |
| abstract_inverted_index.rather | 312 |
| abstract_inverted_index.severe | 411 |
| abstract_inverted_index.strand | 421 |
| abstract_inverted_index.study. | 258 |
| abstract_inverted_index.(C-IDR) | 154 |
| abstract_inverted_index.(FMRP), | 146 |
| abstract_inverted_index.(IDRs). | 106 |
| abstract_inverted_index.DNA:RNA | 10 |
| abstract_inverted_index.Fragile | 143, 428 |
| abstract_inverted_index.Protein | 145 |
| abstract_inverted_index.R-loops | 0, 21, 125, 214 |
| abstract_inverted_index.Reader, | 141 |
| abstract_inverted_index.absence | 397 |
| abstract_inverted_index.another | 197 |
| abstract_inverted_index.between | 379 |
| abstract_inverted_index.binding | 70, 161, 324, 384 |
| abstract_inverted_index.biology | 257 |
| abstract_inverted_index.complex | 195 |
| abstract_inverted_index.contain | 99 |
| abstract_inverted_index.disease | 30 |
| abstract_inverted_index.domains | 71, 117, 168 |
| abstract_inverted_index.explore | 240 |
| abstract_inverted_index.genomic | 412 |
| abstract_inverted_index.hybrid, | 11 |
| abstract_inverted_index.modular | 139 |
| abstract_inverted_index.nucleic | 4, 66, 199 |
| abstract_inverted_index.overall | 362 |
| abstract_inverted_index.perform | 22 |
| abstract_inverted_index.precise | 108 |
| abstract_inverted_index.present | 364 |
| abstract_inverted_index.prevent | 73 |
| abstract_inverted_index.process | 124, 220 |
| abstract_inverted_index.provide | 375 |
| abstract_inverted_index.regions | 105 |
| abstract_inverted_index.resolve | 76 |
| abstract_inverted_index.results | 363 |
| abstract_inverted_index.screens | 52 |
| abstract_inverted_index.several | 331 |
| abstract_inverted_index.through | 251, 388 |
| abstract_inverted_index.undergo | 184, 347 |
| abstract_inverted_index.“mere | 315 |
| abstract_inverted_index.(ss)DNA, | 15 |
| abstract_inverted_index.However, | 86 |
| abstract_inverted_index.Previous | 50 |
| abstract_inverted_index.R-loops, | 58, 311 |
| abstract_inverted_index.R-loops. | 85 |
| abstract_inverted_index.Readers, | 286 |
| abstract_inverted_index.analysis | 328 |
| abstract_inverted_index.assembly | 190, 229, 390 |
| abstract_inverted_index.cellular | 35 |
| abstract_inverted_index.composed | 7 |
| abstract_inverted_index.contains | 287 |
| abstract_inverted_index.critical | 23 |
| abstract_inverted_index.defects, | 413 |
| abstract_inverted_index.directly | 308 |
| abstract_inverted_index.enhanced | 224 |
| abstract_inverted_index.enriched | 293 |
| abstract_inverted_index.exciting | 235 |
| abstract_inverted_index.findings | 236 |
| abstract_inverted_index.flexible | 316 |
| abstract_inverted_index.function | 402 |
| abstract_inverted_index.interact | 309 |
| abstract_inverted_index.majority | 280 |
| abstract_inverted_index.modulate | 127 |
| abstract_inverted_index.multiple | 54 |
| abstract_inverted_index.observed | 275, 426 |
| abstract_inverted_index.prompted | 237 |
| abstract_inverted_index.proteins | 55, 63, 97, 250, 332 |
| abstract_inverted_index.recently | 180, 207, 425 |
| abstract_inverted_index.sequence | 264 |
| abstract_inverted_index.studying | 136 |
| abstract_inverted_index.suppress | 212 |
| abstract_inverted_index.trailing | 18, 171 |
| abstract_inverted_index.displaced | 13 |
| abstract_inverted_index.elucidate | 33 |
| abstract_inverted_index.evaluated | 261 |
| abstract_inverted_index.explored. | 134 |
| abstract_inverted_index.functions | 25 |
| abstract_inverted_index.molecular | 109 |
| abstract_inverted_index.overhang. | 20, 173 |
| abstract_inverted_index.predicted | 339 |
| abstract_inverted_index.recognize | 81, 122 |
| abstract_inverted_index.separated | 357 |
| abstract_inverted_index.sequences | 297 |
| abstract_inverted_index.signaling | 129, 249, 387 |
| abstract_inverted_index.stretches | 101 |
| abstract_inverted_index.synergize | 120 |
| abstract_inverted_index.C-terminal | 152 |
| abstract_inverted_index.N-terminal | 166 |
| abstract_inverted_index.Separation | 188 |
| abstract_inverted_index.associated | 354 |
| abstract_inverted_index.biological | 24 |
| abstract_inverted_index.complexity | 296 |
| abstract_inverted_index.connecting | 318 |
| abstract_inverted_index.containing | 64 |
| abstract_inverted_index.discovered | 149 |
| abstract_inverted_index.disordered | 104 |
| abstract_inverted_index.downstream | 386 |
| abstract_inverted_index.especially | 285 |
| abstract_inverted_index.factories. | 233 |
| abstract_inverted_index.formation, | 45 |
| abstract_inverted_index.functional | 377 |
| abstract_inverted_index.functions, | 36 |
| abstract_inverted_index.helicases, | 78 |
| abstract_inverted_index.hypothesis | 367 |
| abstract_inverted_index.identified | 53 |
| abstract_inverted_index.linkers” | 317 |
| abstract_inverted_index.mechanisms | 42, 112 |
| abstract_inverted_index.persistent | 213 |
| abstract_inverted_index.processing | 68, 247 |
| abstract_inverted_index.propensity | 268 |
| abstract_inverted_index.signaling, | 47 |
| abstract_inverted_index.structural | 111 |
| abstract_inverted_index.structure, | 201 |
| abstract_inverted_index.structures | 6 |
| abstract_inverted_index.suggesting | 303 |
| abstract_inverted_index.underlying | 43 |
| abstract_inverted_index.understand | 40 |
| abstract_inverted_index.widespread | 417 |
| abstract_inverted_index.composition | 265 |
| abstract_inverted_index.conditions. | 31 |
| abstract_inverted_index.domains”. | 325 |
| abstract_inverted_index.interacting | 93 |
| abstract_inverted_index.interactome | 336, 373 |
| abstract_inverted_index.interactors | 406 |
| abstract_inverted_index.nucleases), | 79 |
| abstract_inverted_index.organelles. | 359 |
| abstract_inverted_index.potentially | 408 |
| abstract_inverted_index.predominant | 159 |
| abstract_inverted_index.prevalence, | 263 |
| abstract_inverted_index.recognition | 381 |
| abstract_inverted_index.recognizing | 169 |
| abstract_inverted_index.resolution. | 49 |
| abstract_inverted_index.significant | 88 |
| abstract_inverted_index.spontaneous | 185 |
| abstract_inverted_index.Furthermore, | 204, 326 |
| abstract_inverted_index.IDR-enriched | 404 |
| abstract_inverted_index.composition, | 302 |
| abstract_inverted_index.demonstrated | 345 |
| abstract_inverted_index.interactome, | 284 |
| abstract_inverted_index.interactome. | 273 |
| abstract_inverted_index.membraneless | 231, 358 |
| abstract_inverted_index.recognition, | 46 |
| abstract_inverted_index.unexpectedly | 148 |
| abstract_inverted_index.G-quadruplex. | 203 |
| abstract_inverted_index.LLPS-mediated | 392 |
| abstract_inverted_index.Liquid-Liquid | 186 |
| abstract_inverted_index.comprehensive | 253 |
| abstract_inverted_index.computational | 256 |
| abstract_inverted_index.dysregulation | 399 |
| abstract_inverted_index.intrinsically | 103 |
| abstract_inverted_index.membrane-less | 393 |
| abstract_inverted_index.non-canonical | 198 |
| abstract_inverted_index.transcription | 232 |
| abstract_inverted_index.“functional | 320 |
| abstract_inverted_index.Interestingly, | 174 |
| abstract_inverted_index.bioinformatics | 254 |
| abstract_inverted_index.experimentally | 344 |
| abstract_inverted_index.non-canonical, | 2 |
| abstract_inverted_index.three-stranded | 3 |
| abstract_inverted_index.transcription, | 218 |
| abstract_inverted_index.R-loop-mediated | 128, 418 |
| abstract_inverted_index.high-throughput | 51 |
| abstract_inverted_index.patient-derived | 430 |
| abstract_inverted_index.single-stranded | 14 |
| abstract_inverted_index.topoisomerases), | 75 |
| abstract_inverted_index.thought-provoking | 366 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 97 |
| corresponding_author_ids | https://openalex.org/A5086367121 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 10 |
| corresponding_institution_ids | https://openalex.org/I20388574 |
| citation_normalized_percentile.value | 0.89385285 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |