Abstract 126: Multidimensional gene expression models for characterizing response and metastasis in solid tumor samples Article Swipe
 YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1158/1538-7445.sabcs18-126
YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1158/1538-7445.sabcs18-126
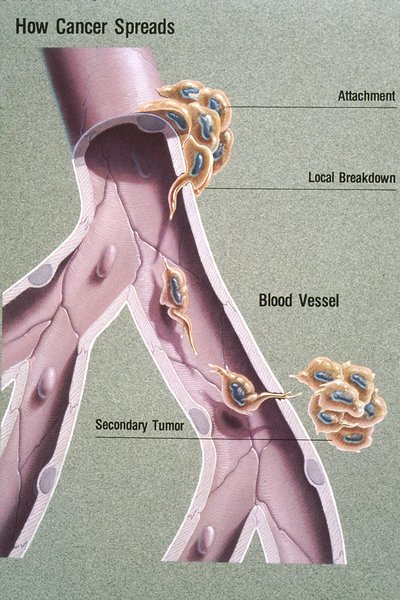
Predicting response to standard of care therapies and new immunotherapies is enabled by tools that quantitatively analyze the complex heterogeneity of the tumor microenvironment. Current methods to do so primarily rely either on single gene or protein markers, and often report solely on one feature of the multifaceted immune cycle. Unlike other molecular approaches, the ImmunoPrismTM Immune Profiling Assay has been built using multidimensional gene expression models validated to accurately identify immune cells in a complex mixture with high correlation to flow cytometry measurements, but accessible for formalin-fixed paraffin-embedded (FFPE) tissues. We will report results showing the assay is highly sensitive, capable of detecting immune cells present to as little as 2% of the specimen, important for critical immune cell types such as M1 and M2 macrophages and Tregs. Further, the assay requires as few as two sections of FFPE solid tumor tissue or as little as 20 ng of total RNA which enables the analysis of rare clinical archives. In this study, we apply the ImmunoPrism assay to cohorts of patients with indications including non-small cell lung carcinoma (NSCLC), triple-negative breast cancer (TNBC) and pancreatic adenocarcinoma (ADC) treated with standard of care therapies. Immune profile changes for pre vs. post treatment and primary vs. metastatic tissue are reported in the context of clinical data, such as therapy response. Resulting biomarkers, including a novel machine-learning based multidimensional marker are also reported with predictive accuracy, PPV, NPV, and selection thresholds.Citation Format: Walter P. Carney, Milan Bhagat, Natalie LaFranzo. Multidimensional gene expression models for characterizing response and metastasis in solid tumor samples [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2019; 2019 Mar 29-Apr 3; Atlanta, GA. Philadelphia (PA): AACR; Cancer Res 2019;79(13 Suppl):Abstract nr 126.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1158/1538-7445.sabcs18-126
- OA Status
- gold
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4246634663
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4246634663Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1158/1538-7445.sabcs18-126Digital Object Identifier
- Title
-
Abstract 126: Multidimensional gene expression models for characterizing response and metastasis in solid tumor samplesWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2019Year of publication
- Publication date
-
2019-07-01Full publication date if available
- Authors
-
Walter P. Carney, Milan Bhagat, Natalie A. LaFranzoList of authors in order
- Landing page
-
https://doi.org/10.1158/1538-7445.sabcs18-126Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1158/1538-7445.sabcs18-126Direct OA link when available
- Concepts
-
Immune system, Tumor microenvironment, Context (archaeology), Flow cytometry, Cancer research, Breast cancer, Gene expression, Adenocarcinoma, Gene expression profiling, Computational biology, Metastasis, Cancer, Biology, Medicine, Gene, Immunology, Internal medicine, Biochemistry, PaleontologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4246634663 |
|---|---|
| doi | https://doi.org/10.1158/1538-7445.sabcs18-126 |
| ids.doi | https://doi.org/10.1158/1538-7445.sabcs18-126 |
| ids.openalex | https://openalex.org/W4246634663 |
| fwci | 0.0 |
| type | article |
| title | Abstract 126: Multidimensional gene expression models for characterizing response and metastasis in solid tumor samples |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | 126 |
| biblio.first_page | 126 |
| topics[0].id | https://openalex.org/T10158 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.973800003528595 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2730 |
| topics[0].subfield.display_name | Oncology |
| topics[0].display_name | Cancer Immunotherapy and Biomarkers |
| topics[1].id | https://openalex.org/T12422 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.963100016117096 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2741 |
| topics[1].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[1].display_name | Radiomics and Machine Learning in Medical Imaging |
| topics[2].id | https://openalex.org/T11297 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9517999887466431 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2740 |
| topics[2].subfield.display_name | Pulmonary and Respiratory Medicine |
| topics[2].display_name | Ferroptosis and cancer prognosis |
| is_xpac | False |
| apc_list.value | 2000 |
| apc_list.currency | USD |
| apc_list.value_usd | 2000 |
| apc_paid.value | 2000 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2000 |
| concepts[0].id | https://openalex.org/C8891405 |
| concepts[0].level | 2 |
| concepts[0].score | 0.6717486381530762 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1059 |
| concepts[0].display_name | Immune system |
| concepts[1].id | https://openalex.org/C2776107976 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5444245934486389 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1786433 |
| concepts[1].display_name | Tumor microenvironment |
| concepts[2].id | https://openalex.org/C2779343474 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5187606811523438 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q3109175 |
| concepts[2].display_name | Context (archaeology) |
| concepts[3].id | https://openalex.org/C553184892 |
| concepts[3].level | 2 |
| concepts[3].score | 0.4980161190032959 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1141429 |
| concepts[3].display_name | Flow cytometry |
| concepts[4].id | https://openalex.org/C502942594 |
| concepts[4].level | 1 |
| concepts[4].score | 0.4916685223579407 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q3421914 |
| concepts[4].display_name | Cancer research |
| concepts[5].id | https://openalex.org/C530470458 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4808819591999054 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q128581 |
| concepts[5].display_name | Breast cancer |
| concepts[6].id | https://openalex.org/C150194340 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4517766833305359 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[6].display_name | Gene expression |
| concepts[7].id | https://openalex.org/C2781182431 |
| concepts[7].level | 3 |
| concepts[7].score | 0.4362603425979614 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q356033 |
| concepts[7].display_name | Adenocarcinoma |
| concepts[8].id | https://openalex.org/C18431079 |
| concepts[8].level | 4 |
| concepts[8].score | 0.4257246255874634 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q1502169 |
| concepts[8].display_name | Gene expression profiling |
| concepts[9].id | https://openalex.org/C70721500 |
| concepts[9].level | 1 |
| concepts[9].score | 0.42525020241737366 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[9].display_name | Computational biology |
| concepts[10].id | https://openalex.org/C2779013556 |
| concepts[10].level | 3 |
| concepts[10].score | 0.4185592830181122 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q181876 |
| concepts[10].display_name | Metastasis |
| concepts[11].id | https://openalex.org/C121608353 |
| concepts[11].level | 2 |
| concepts[11].score | 0.4050099551677704 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[11].display_name | Cancer |
| concepts[12].id | https://openalex.org/C86803240 |
| concepts[12].level | 0 |
| concepts[12].score | 0.3864212930202484 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[12].display_name | Biology |
| concepts[13].id | https://openalex.org/C71924100 |
| concepts[13].level | 0 |
| concepts[13].score | 0.37482312321662903 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[13].display_name | Medicine |
| concepts[14].id | https://openalex.org/C104317684 |
| concepts[14].level | 2 |
| concepts[14].score | 0.25466424226760864 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[14].display_name | Gene |
| concepts[15].id | https://openalex.org/C203014093 |
| concepts[15].level | 1 |
| concepts[15].score | 0.23237106204032898 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[15].display_name | Immunology |
| concepts[16].id | https://openalex.org/C126322002 |
| concepts[16].level | 1 |
| concepts[16].score | 0.2151281237602234 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[16].display_name | Internal medicine |
| concepts[17].id | https://openalex.org/C55493867 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[17].display_name | Biochemistry |
| concepts[18].id | https://openalex.org/C151730666 |
| concepts[18].level | 1 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q7205 |
| concepts[18].display_name | Paleontology |
| keywords[0].id | https://openalex.org/keywords/immune-system |
| keywords[0].score | 0.6717486381530762 |
| keywords[0].display_name | Immune system |
| keywords[1].id | https://openalex.org/keywords/tumor-microenvironment |
| keywords[1].score | 0.5444245934486389 |
| keywords[1].display_name | Tumor microenvironment |
| keywords[2].id | https://openalex.org/keywords/context |
| keywords[2].score | 0.5187606811523438 |
| keywords[2].display_name | Context (archaeology) |
| keywords[3].id | https://openalex.org/keywords/flow-cytometry |
| keywords[3].score | 0.4980161190032959 |
| keywords[3].display_name | Flow cytometry |
| keywords[4].id | https://openalex.org/keywords/cancer-research |
| keywords[4].score | 0.4916685223579407 |
| keywords[4].display_name | Cancer research |
| keywords[5].id | https://openalex.org/keywords/breast-cancer |
| keywords[5].score | 0.4808819591999054 |
| keywords[5].display_name | Breast cancer |
| keywords[6].id | https://openalex.org/keywords/gene-expression |
| keywords[6].score | 0.4517766833305359 |
| keywords[6].display_name | Gene expression |
| keywords[7].id | https://openalex.org/keywords/adenocarcinoma |
| keywords[7].score | 0.4362603425979614 |
| keywords[7].display_name | Adenocarcinoma |
| keywords[8].id | https://openalex.org/keywords/gene-expression-profiling |
| keywords[8].score | 0.4257246255874634 |
| keywords[8].display_name | Gene expression profiling |
| keywords[9].id | https://openalex.org/keywords/computational-biology |
| keywords[9].score | 0.42525020241737366 |
| keywords[9].display_name | Computational biology |
| keywords[10].id | https://openalex.org/keywords/metastasis |
| keywords[10].score | 0.4185592830181122 |
| keywords[10].display_name | Metastasis |
| keywords[11].id | https://openalex.org/keywords/cancer |
| keywords[11].score | 0.4050099551677704 |
| keywords[11].display_name | Cancer |
| keywords[12].id | https://openalex.org/keywords/biology |
| keywords[12].score | 0.3864212930202484 |
| keywords[12].display_name | Biology |
| keywords[13].id | https://openalex.org/keywords/medicine |
| keywords[13].score | 0.37482312321662903 |
| keywords[13].display_name | Medicine |
| keywords[14].id | https://openalex.org/keywords/gene |
| keywords[14].score | 0.25466424226760864 |
| keywords[14].display_name | Gene |
| keywords[15].id | https://openalex.org/keywords/immunology |
| keywords[15].score | 0.23237106204032898 |
| keywords[15].display_name | Immunology |
| keywords[16].id | https://openalex.org/keywords/internal-medicine |
| keywords[16].score | 0.2151281237602234 |
| keywords[16].display_name | Internal medicine |
| language | en |
| locations[0].id | doi:10.1158/1538-7445.sabcs18-126 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S12644804 |
| locations[0].source.issn | 1010-4283, 1423-0380 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1010-4283 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Tumor Biology |
| locations[0].source.host_organization | https://openalex.org/P4310320017 |
| locations[0].source.host_organization_name | SAGE Publishing |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320017 |
| locations[0].source.host_organization_lineage_names | SAGE Publishing |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | proceedings-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Tumor Biology |
| locations[0].landing_page_url | https://doi.org/10.1158/1538-7445.sabcs18-126 |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5051230211 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Walter P. Carney |
| authorships[0].affiliations[0].raw_affiliation_string | TristarGroup, LLC, Washington D.C., WA |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Walter P. Carney |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | TristarGroup, LLC, Washington D.C., WA |
| authorships[1].author.id | https://openalex.org/A5060619682 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Milan Bhagat |
| authorships[1].affiliations[0].raw_affiliation_string | TristarGroup, LLC, Washington D.C., WA |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Milan Bhagat |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | TristarGroup, LLC, Washington D.C., WA |
| authorships[2].author.id | https://openalex.org/A5018540261 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-1009-4533 |
| authorships[2].author.display_name | Natalie A. LaFranzo |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210127773 |
| authorships[2].affiliations[0].raw_affiliation_string | Cofactor Genomics, St Louis, MO. |
| authorships[2].institutions[0].id | https://openalex.org/I4210127773 |
| authorships[2].institutions[0].ror | https://ror.org/031fg5f22 |
| authorships[2].institutions[0].type | company |
| authorships[2].institutions[0].lineage | https://openalex.org/I4210127773 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Cofactor Genomics (United States) |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Natalie LaFranzo |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Cofactor Genomics, St Louis, MO. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1158/1538-7445.sabcs18-126 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Abstract 126: Multidimensional gene expression models for characterizing response and metastasis in solid tumor samples |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10158 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.973800003528595 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2730 |
| primary_topic.subfield.display_name | Oncology |
| primary_topic.display_name | Cancer Immunotherapy and Biomarkers |
| related_works | https://openalex.org/W2500038226, https://openalex.org/W1966652204, https://openalex.org/W4307869277, https://openalex.org/W2415281480, https://openalex.org/W4251194061, https://openalex.org/W2314330891, https://openalex.org/W4312063994, https://openalex.org/W2052058314, https://openalex.org/W2018894105, https://openalex.org/W3109885192 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1158/1538-7445.sabcs18-126 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S12644804 |
| best_oa_location.source.issn | 1010-4283, 1423-0380 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1010-4283 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Tumor Biology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320017 |
| best_oa_location.source.host_organization_name | SAGE Publishing |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320017 |
| best_oa_location.source.host_organization_lineage_names | SAGE Publishing |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | proceedings-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Tumor Biology |
| best_oa_location.landing_page_url | https://doi.org/10.1158/1538-7445.sabcs18-126 |
| primary_location.id | doi:10.1158/1538-7445.sabcs18-126 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S12644804 |
| primary_location.source.issn | 1010-4283, 1423-0380 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1010-4283 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Tumor Biology |
| primary_location.source.host_organization | https://openalex.org/P4310320017 |
| primary_location.source.host_organization_name | SAGE Publishing |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320017 |
| primary_location.source.host_organization_lineage_names | SAGE Publishing |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | proceedings-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Tumor Biology |
| primary_location.landing_page_url | https://doi.org/10.1158/1538-7445.sabcs18-126 |
| publication_date | 2019-07-01 |
| publication_year | 2019 |
| referenced_works_count | 0 |
| abstract_inverted_index.a | 74, 222 |
| abstract_inverted_index.2% | 111 |
| abstract_inverted_index.20 | 147 |
| abstract_inverted_index.3; | 276 |
| abstract_inverted_index.In | 160 |
| abstract_inverted_index.M1 | 123 |
| abstract_inverted_index.M2 | 125 |
| abstract_inverted_index.P. | 241 |
| abstract_inverted_index.We | 91 |
| abstract_inverted_index.as | 108, 110, 122, 133, 135, 144, 146, 216 |
| abstract_inverted_index.by | 12 |
| abstract_inverted_index.do | 27 |
| abstract_inverted_index.in | 73, 209, 256 |
| abstract_inverted_index.is | 10, 98 |
| abstract_inverted_index.ng | 148 |
| abstract_inverted_index.nr | 286 |
| abstract_inverted_index.of | 4, 20, 45, 102, 112, 138, 149, 156, 170, 191, 212, 263 |
| abstract_inverted_index.on | 32, 42 |
| abstract_inverted_index.or | 35, 143 |
| abstract_inverted_index.so | 28 |
| abstract_inverted_index.to | 2, 26, 68, 80, 107, 168 |
| abstract_inverted_index.we | 163 |
| abstract_inverted_index.GA. | 278 |
| abstract_inverted_index.In: | 261 |
| abstract_inverted_index.Mar | 274 |
| abstract_inverted_index.RNA | 151 |
| abstract_inverted_index.Res | 283 |
| abstract_inverted_index.and | 7, 38, 124, 127, 184, 202, 236, 254 |
| abstract_inverted_index.are | 207, 228 |
| abstract_inverted_index.but | 84 |
| abstract_inverted_index.few | 134 |
| abstract_inverted_index.for | 86, 116, 197, 251, 267 |
| abstract_inverted_index.has | 59 |
| abstract_inverted_index.new | 8 |
| abstract_inverted_index.one | 43 |
| abstract_inverted_index.pre | 198 |
| abstract_inverted_index.the | 17, 21, 46, 54, 96, 113, 130, 154, 165, 210, 264 |
| abstract_inverted_index.two | 136 |
| abstract_inverted_index.vs. | 199, 204 |
| abstract_inverted_index.126. | 287 |
| abstract_inverted_index.2019 | 273 |
| abstract_inverted_index.FFPE | 139 |
| abstract_inverted_index.NPV, | 235 |
| abstract_inverted_index.PPV, | 234 |
| abstract_inverted_index.also | 229 |
| abstract_inverted_index.been | 60 |
| abstract_inverted_index.care | 5, 192 |
| abstract_inverted_index.cell | 119, 176 |
| abstract_inverted_index.flow | 81 |
| abstract_inverted_index.gene | 34, 64, 248 |
| abstract_inverted_index.high | 78 |
| abstract_inverted_index.lung | 177 |
| abstract_inverted_index.post | 200 |
| abstract_inverted_index.rare | 157 |
| abstract_inverted_index.rely | 30 |
| abstract_inverted_index.such | 121, 215 |
| abstract_inverted_index.that | 14 |
| abstract_inverted_index.this | 161 |
| abstract_inverted_index.will | 92 |
| abstract_inverted_index.with | 77, 172, 189, 231 |
| abstract_inverted_index.(ADC) | 187 |
| abstract_inverted_index.(PA): | 280 |
| abstract_inverted_index.2019; | 272 |
| abstract_inverted_index.AACR; | 281 |
| abstract_inverted_index.Assay | 58 |
| abstract_inverted_index.Milan | 243 |
| abstract_inverted_index.apply | 164 |
| abstract_inverted_index.assay | 97, 131, 167 |
| abstract_inverted_index.based | 225 |
| abstract_inverted_index.built | 61 |
| abstract_inverted_index.cells | 72, 105 |
| abstract_inverted_index.data, | 214 |
| abstract_inverted_index.novel | 223 |
| abstract_inverted_index.often | 39 |
| abstract_inverted_index.other | 51 |
| abstract_inverted_index.solid | 140, 257 |
| abstract_inverted_index.tools | 13 |
| abstract_inverted_index.total | 150 |
| abstract_inverted_index.tumor | 22, 141, 258 |
| abstract_inverted_index.types | 120 |
| abstract_inverted_index.using | 62 |
| abstract_inverted_index.which | 152 |
| abstract_inverted_index.(FFPE) | 89 |
| abstract_inverted_index.(TNBC) | 183 |
| abstract_inverted_index.29-Apr | 275 |
| abstract_inverted_index.Annual | 270 |
| abstract_inverted_index.Cancer | 268, 282 |
| abstract_inverted_index.Immune | 56, 194 |
| abstract_inverted_index.Tregs. | 128 |
| abstract_inverted_index.Unlike | 50 |
| abstract_inverted_index.Walter | 240 |
| abstract_inverted_index.breast | 181 |
| abstract_inverted_index.cancer | 182 |
| abstract_inverted_index.cycle. | 49 |
| abstract_inverted_index.either | 31 |
| abstract_inverted_index.highly | 99 |
| abstract_inverted_index.immune | 48, 71, 104, 118 |
| abstract_inverted_index.little | 109, 145 |
| abstract_inverted_index.marker | 227 |
| abstract_inverted_index.models | 66, 250 |
| abstract_inverted_index.report | 40, 93 |
| abstract_inverted_index.single | 33 |
| abstract_inverted_index.solely | 41 |
| abstract_inverted_index.study, | 162 |
| abstract_inverted_index.tissue | 142, 206 |
| abstract_inverted_index.Bhagat, | 244 |
| abstract_inverted_index.Carney, | 242 |
| abstract_inverted_index.Current | 24 |
| abstract_inverted_index.Format: | 239 |
| abstract_inverted_index.Meeting | 271 |
| abstract_inverted_index.Natalie | 245 |
| abstract_inverted_index.analyze | 16 |
| abstract_inverted_index.capable | 101 |
| abstract_inverted_index.changes | 196 |
| abstract_inverted_index.cohorts | 169 |
| abstract_inverted_index.complex | 18, 75 |
| abstract_inverted_index.context | 211 |
| abstract_inverted_index.enabled | 11 |
| abstract_inverted_index.enables | 153 |
| abstract_inverted_index.feature | 44 |
| abstract_inverted_index.methods | 25 |
| abstract_inverted_index.mixture | 76 |
| abstract_inverted_index.present | 106 |
| abstract_inverted_index.primary | 203 |
| abstract_inverted_index.profile | 195 |
| abstract_inverted_index.protein | 36 |
| abstract_inverted_index.results | 94 |
| abstract_inverted_index.samples | 259 |
| abstract_inverted_index.showing | 95 |
| abstract_inverted_index.therapy | 217 |
| abstract_inverted_index.treated | 188 |
| abstract_inverted_index.(NSCLC), | 179 |
| abstract_inverted_index.American | 265 |
| abstract_inverted_index.Atlanta, | 277 |
| abstract_inverted_index.Further, | 129 |
| abstract_inverted_index.Research | 269 |
| abstract_inverted_index.analysis | 155 |
| abstract_inverted_index.clinical | 158, 213 |
| abstract_inverted_index.critical | 117 |
| abstract_inverted_index.identify | 70 |
| abstract_inverted_index.markers, | 37 |
| abstract_inverted_index.patients | 171 |
| abstract_inverted_index.reported | 208, 230 |
| abstract_inverted_index.requires | 132 |
| abstract_inverted_index.response | 1, 253 |
| abstract_inverted_index.sections | 137 |
| abstract_inverted_index.standard | 3, 190 |
| abstract_inverted_index.tissues. | 90 |
| abstract_inverted_index.LaFranzo. | 246 |
| abstract_inverted_index.Profiling | 57 |
| abstract_inverted_index.Resulting | 219 |
| abstract_inverted_index.accuracy, | 233 |
| abstract_inverted_index.archives. | 159 |
| abstract_inverted_index.carcinoma | 178 |
| abstract_inverted_index.cytometry | 82 |
| abstract_inverted_index.detecting | 103 |
| abstract_inverted_index.important | 115 |
| abstract_inverted_index.including | 174, 221 |
| abstract_inverted_index.molecular | 52 |
| abstract_inverted_index.non-small | 175 |
| abstract_inverted_index.primarily | 29 |
| abstract_inverted_index.response. | 218 |
| abstract_inverted_index.selection | 237 |
| abstract_inverted_index.specimen, | 114 |
| abstract_inverted_index.therapies | 6 |
| abstract_inverted_index.treatment | 201 |
| abstract_inverted_index.validated | 67 |
| abstract_inverted_index.2019;79(13 | 284 |
| abstract_inverted_index.Predicting | 0 |
| abstract_inverted_index.accessible | 85 |
| abstract_inverted_index.accurately | 69 |
| abstract_inverted_index.expression | 65, 249 |
| abstract_inverted_index.metastasis | 255 |
| abstract_inverted_index.metastatic | 205 |
| abstract_inverted_index.pancreatic | 185 |
| abstract_inverted_index.predictive | 232 |
| abstract_inverted_index.sensitive, | 100 |
| abstract_inverted_index.therapies. | 193 |
| abstract_inverted_index.Association | 266 |
| abstract_inverted_index.ImmunoPrism | 166 |
| abstract_inverted_index.Proceedings | 262 |
| abstract_inverted_index.[abstract]. | 260 |
| abstract_inverted_index.approaches, | 53 |
| abstract_inverted_index.biomarkers, | 220 |
| abstract_inverted_index.correlation | 79 |
| abstract_inverted_index.indications | 173 |
| abstract_inverted_index.macrophages | 126 |
| abstract_inverted_index.Philadelphia | 279 |
| abstract_inverted_index.multifaceted | 47 |
| abstract_inverted_index.ImmunoPrismTM | 55 |
| abstract_inverted_index.heterogeneity | 19 |
| abstract_inverted_index.measurements, | 83 |
| abstract_inverted_index.adenocarcinoma | 186 |
| abstract_inverted_index.characterizing | 252 |
| abstract_inverted_index.formalin-fixed | 87 |
| abstract_inverted_index.quantitatively | 15 |
| abstract_inverted_index.Suppl):Abstract | 285 |
| abstract_inverted_index.immunotherapies | 9 |
| abstract_inverted_index.triple-negative | 180 |
| abstract_inverted_index.Multidimensional | 247 |
| abstract_inverted_index.machine-learning | 224 |
| abstract_inverted_index.multidimensional | 63, 226 |
| abstract_inverted_index.microenvironment. | 23 |
| abstract_inverted_index.paraffin-embedded | 88 |
| abstract_inverted_index.thresholds.Citation | 238 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/2 |
| sustainable_development_goals[0].score | 0.5600000023841858 |
| sustainable_development_goals[0].display_name | Zero hunger |
| citation_normalized_percentile.value | 0.34639671 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |