Additional file 1 of Evaluation of CRISPR gene-editing tools in zebrafish Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.6084/m9.figshare.17913224
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.6084/m9.figshare.17913224
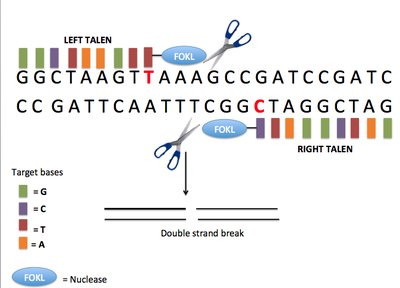
Additional file 1: Supplementary Figure 1. Editing efficiencies across 50 gRNAs with different GC content. Supplementary Figure 2. Mosaicism predicted using Illumina sequencing of G0 mutant larvae. Supplementary Figure 3. Network analyses of the shared differentially expressed genes in Cas9 enzyme and Cas9 mRNA injected larvae. Supplementary Table 1. Oligonucleotides utilized for all assays performed throughout the project. Supplementary Table 2. On-target efficiency scores for the 50 gRNAs obtained from different in silico prediction tools, empirical cleavage scores in vivo using Sanger sequencing and sequence deconvolution tools (TIDE and ICE), from the in vitro assay CIRCLE-seq (RPMN), and Illumina sequencing using the CrispRVariants software. Supplementary Table 3. Description of the off-targets predicted by CRISPRScan and CIRCLE-seq for the 50 evaluated gRNAs. The percentage of sites intersecting genes from the CRISPRScan tool was obtained from the top 30 predicted off-targets since the tool only provides detailed information from these. Supplementary Table 4. List of the frameshift variants identified in protein-coding genes in samples injected with either Cas9 enzyme or Cas9 mRNA using uninjected batch-siblings as reference. Supplementary Table 5. Illumina mosaicism percentages observed in larvae injected with Cas9 (enzyme or mRNA), catalytically dead Cas9 (dCas9), scrambled gRNA, uninjected batch siblings, and a finclip from their crossing parents. Supplementary Table 6. List of DE genes observed in Cas9-injected samples relative to uninjected batch-siblings and siblings from another batch. Supplementary Table 7. List of gene ontology terms enriched in upregulated genes found in both injection treatments (Cas9 enzyme and Cas9 mRNA).
Related Topics
- Type
- dataset
- Language
- en
- Landing Page
- https://doi.org/10.6084/m9.figshare.17913224
- OA Status
- gold
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4394277886
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4394277886Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.6084/m9.figshare.17913224Digital Object Identifier
- Title
-
Additional file 1 of Evaluation of CRISPR gene-editing tools in zebrafishWork title
- Type
-
datasetOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-01-01Full publication date if available
- Authors
-
José M. Uribe‐Salazar, Gulhan Kaya, Aadithya Sekar, KaeChandra Weyenberg, Cole Ingamells, Megan Y. DennisList of authors in order
- Landing page
-
https://doi.org/10.6084/m9.figshare.17913224Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.6084/m9.figshare.17913224Direct OA link when available
- Concepts
-
CRISPR, Zebrafish, Genome editing, Computational biology, Computer science, Gene, Biology, GeneticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4394277886 |
|---|---|
| doi | https://doi.org/10.6084/m9.figshare.17913224 |
| ids.doi | https://doi.org/10.6084/m9.figshare.17913224 |
| ids.openalex | https://openalex.org/W4394277886 |
| fwci | |
| type | dataset |
| title | Additional file 1 of Evaluation of CRISPR gene-editing tools in zebrafish |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10878 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9965999722480774 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | CRISPR and Genetic Engineering |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C98108389 |
| concepts[0].level | 3 |
| concepts[0].score | 0.9259204864501953 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q412563 |
| concepts[0].display_name | CRISPR |
| concepts[1].id | https://openalex.org/C2776878037 |
| concepts[1].level | 3 |
| concepts[1].score | 0.8090776205062866 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q169444 |
| concepts[1].display_name | Zebrafish |
| concepts[2].id | https://openalex.org/C144501496 |
| concepts[2].level | 4 |
| concepts[2].score | 0.7231947183609009 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q5533489 |
| concepts[2].display_name | Genome editing |
| concepts[3].id | https://openalex.org/C70721500 |
| concepts[3].level | 1 |
| concepts[3].score | 0.5370371341705322 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[3].display_name | Computational biology |
| concepts[4].id | https://openalex.org/C41008148 |
| concepts[4].level | 0 |
| concepts[4].score | 0.5111420750617981 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[4].display_name | Computer science |
| concepts[5].id | https://openalex.org/C104317684 |
| concepts[5].level | 2 |
| concepts[5].score | 0.3960627019405365 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[5].display_name | Gene |
| concepts[6].id | https://openalex.org/C86803240 |
| concepts[6].level | 0 |
| concepts[6].score | 0.3886456787586212 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[6].display_name | Biology |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.29082387685775757 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| keywords[0].id | https://openalex.org/keywords/crispr |
| keywords[0].score | 0.9259204864501953 |
| keywords[0].display_name | CRISPR |
| keywords[1].id | https://openalex.org/keywords/zebrafish |
| keywords[1].score | 0.8090776205062866 |
| keywords[1].display_name | Zebrafish |
| keywords[2].id | https://openalex.org/keywords/genome-editing |
| keywords[2].score | 0.7231947183609009 |
| keywords[2].display_name | Genome editing |
| keywords[3].id | https://openalex.org/keywords/computational-biology |
| keywords[3].score | 0.5370371341705322 |
| keywords[3].display_name | Computational biology |
| keywords[4].id | https://openalex.org/keywords/computer-science |
| keywords[4].score | 0.5111420750617981 |
| keywords[4].display_name | Computer science |
| keywords[5].id | https://openalex.org/keywords/gene |
| keywords[5].score | 0.3960627019405365 |
| keywords[5].display_name | Gene |
| keywords[6].id | https://openalex.org/keywords/biology |
| keywords[6].score | 0.3886456787586212 |
| keywords[6].display_name | Biology |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.29082387685775757 |
| keywords[7].display_name | Genetics |
| language | en |
| locations[0].id | doi:10.6084/m9.figshare.17913224 |
| locations[0].is_oa | True |
| locations[0].source | |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | dataset |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.6084/m9.figshare.17913224 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5011005586 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-4875-818X |
| authorships[0].author.display_name | José M. Uribe‐Salazar |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | José M. Uribe-Salazar |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5009295217 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3142-7886 |
| authorships[1].author.display_name | Gulhan Kaya |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Gulhan Kaya |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5059426994 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-1503-2529 |
| authorships[2].author.display_name | Aadithya Sekar |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Aadithya Sekar |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5022460769 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-3354-6280 |
| authorships[3].author.display_name | KaeChandra Weyenberg |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | KaeChandra Weyenberg |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5056409439 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-4706-2666 |
| authorships[4].author.display_name | Cole Ingamells |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Cole Ingamells |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5074379383 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-8502-5420 |
| authorships[5].author.display_name | Megan Y. Dennis |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | Megan Y. Dennis |
| authorships[5].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.6084/m9.figshare.17913224 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Additional file 1 of Evaluation of CRISPR gene-editing tools in zebrafish |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T10878 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9965999722480774 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | CRISPR and Genetic Engineering |
| related_works | https://openalex.org/W4366822718, https://openalex.org/W4200005163, https://openalex.org/W3030388918, https://openalex.org/W2807464733, https://openalex.org/W4220766053, https://openalex.org/W2603161682, https://openalex.org/W4213248511, https://openalex.org/W2991257722, https://openalex.org/W3165203721, https://openalex.org/W2960163646 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.6084/m9.figshare.17913224 |
| best_oa_location.is_oa | True |
| best_oa_location.source | |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | dataset |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.6084/m9.figshare.17913224 |
| primary_location.id | doi:10.6084/m9.figshare.17913224 |
| primary_location.is_oa | True |
| primary_location.source | |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | dataset |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.6084/m9.figshare.17913224 |
| publication_date | 2022-01-01 |
| publication_year | 2022 |
| referenced_works_count | 0 |
| abstract_inverted_index.a | 200 |
| abstract_inverted_index.1. | 5, 48 |
| abstract_inverted_index.1: | 2 |
| abstract_inverted_index.2. | 17, 60 |
| abstract_inverted_index.3. | 29, 106 |
| abstract_inverted_index.30 | 136 |
| abstract_inverted_index.4. | 150 |
| abstract_inverted_index.5. | 177 |
| abstract_inverted_index.50 | 9, 66, 118 |
| abstract_inverted_index.6. | 208 |
| abstract_inverted_index.7. | 228 |
| abstract_inverted_index.DE | 211 |
| abstract_inverted_index.G0 | 24 |
| abstract_inverted_index.GC | 13 |
| abstract_inverted_index.as | 173 |
| abstract_inverted_index.by | 112 |
| abstract_inverted_index.in | 38, 71, 78, 92, 157, 160, 182, 214, 235, 239 |
| abstract_inverted_index.of | 23, 32, 108, 123, 152, 210, 230 |
| abstract_inverted_index.or | 167, 188 |
| abstract_inverted_index.to | 218 |
| abstract_inverted_index.The | 121 |
| abstract_inverted_index.all | 52 |
| abstract_inverted_index.and | 41, 83, 88, 97, 114, 199, 221, 245 |
| abstract_inverted_index.for | 51, 64, 116 |
| abstract_inverted_index.the | 33, 56, 65, 91, 101, 109, 117, 128, 134, 140, 153 |
| abstract_inverted_index.top | 135 |
| abstract_inverted_index.was | 131 |
| abstract_inverted_index.Cas9 | 39, 42, 165, 168, 186, 192, 246 |
| abstract_inverted_index.List | 151, 209, 229 |
| abstract_inverted_index.both | 240 |
| abstract_inverted_index.dead | 191 |
| abstract_inverted_index.file | 1 |
| abstract_inverted_index.from | 69, 90, 127, 133, 146, 202, 223 |
| abstract_inverted_index.gene | 231 |
| abstract_inverted_index.mRNA | 43, 169 |
| abstract_inverted_index.only | 142 |
| abstract_inverted_index.tool | 130, 141 |
| abstract_inverted_index.vivo | 79 |
| abstract_inverted_index.with | 11, 163, 185 |
| abstract_inverted_index.(Cas9 | 243 |
| abstract_inverted_index.(TIDE | 87 |
| abstract_inverted_index.ICE), | 89 |
| abstract_inverted_index.Table | 47, 59, 105, 149, 176, 207, 227 |
| abstract_inverted_index.assay | 94 |
| abstract_inverted_index.batch | 197 |
| abstract_inverted_index.found | 238 |
| abstract_inverted_index.gRNA, | 195 |
| abstract_inverted_index.gRNAs | 10, 67 |
| abstract_inverted_index.genes | 37, 126, 159, 212, 237 |
| abstract_inverted_index.since | 139 |
| abstract_inverted_index.sites | 124 |
| abstract_inverted_index.terms | 233 |
| abstract_inverted_index.their | 203 |
| abstract_inverted_index.tools | 86 |
| abstract_inverted_index.using | 20, 80, 100, 170 |
| abstract_inverted_index.vitro | 93 |
| abstract_inverted_index.Figure | 4, 16, 28 |
| abstract_inverted_index.Sanger | 81 |
| abstract_inverted_index.across | 8 |
| abstract_inverted_index.assays | 53 |
| abstract_inverted_index.batch. | 225 |
| abstract_inverted_index.either | 164 |
| abstract_inverted_index.enzyme | 40, 166, 244 |
| abstract_inverted_index.gRNAs. | 120 |
| abstract_inverted_index.larvae | 183 |
| abstract_inverted_index.mRNA), | 189 |
| abstract_inverted_index.mRNA). | 247 |
| abstract_inverted_index.mutant | 25 |
| abstract_inverted_index.scores | 63, 77 |
| abstract_inverted_index.shared | 34 |
| abstract_inverted_index.silico | 72 |
| abstract_inverted_index.these. | 147 |
| abstract_inverted_index.tools, | 74 |
| abstract_inverted_index.(RPMN), | 96 |
| abstract_inverted_index.(enzyme | 187 |
| abstract_inverted_index.Editing | 6 |
| abstract_inverted_index.Network | 30 |
| abstract_inverted_index.another | 224 |
| abstract_inverted_index.finclip | 201 |
| abstract_inverted_index.larvae. | 26, 45 |
| abstract_inverted_index.samples | 161, 216 |
| abstract_inverted_index.(dCas9), | 193 |
| abstract_inverted_index.Illumina | 21, 98, 178 |
| abstract_inverted_index.analyses | 31 |
| abstract_inverted_index.cleavage | 76 |
| abstract_inverted_index.content. | 14 |
| abstract_inverted_index.crossing | 204 |
| abstract_inverted_index.detailed | 144 |
| abstract_inverted_index.enriched | 234 |
| abstract_inverted_index.injected | 44, 162, 184 |
| abstract_inverted_index.observed | 181, 213 |
| abstract_inverted_index.obtained | 68, 132 |
| abstract_inverted_index.ontology | 232 |
| abstract_inverted_index.parents. | 205 |
| abstract_inverted_index.project. | 57 |
| abstract_inverted_index.provides | 143 |
| abstract_inverted_index.relative | 217 |
| abstract_inverted_index.sequence | 84 |
| abstract_inverted_index.siblings | 222 |
| abstract_inverted_index.utilized | 50 |
| abstract_inverted_index.variants | 155 |
| abstract_inverted_index.Mosaicism | 18 |
| abstract_inverted_index.On-target | 61 |
| abstract_inverted_index.different | 12, 70 |
| abstract_inverted_index.empirical | 75 |
| abstract_inverted_index.evaluated | 119 |
| abstract_inverted_index.expressed | 36 |
| abstract_inverted_index.injection | 241 |
| abstract_inverted_index.mosaicism | 179 |
| abstract_inverted_index.performed | 54 |
| abstract_inverted_index.predicted | 19, 111, 137 |
| abstract_inverted_index.scrambled | 194 |
| abstract_inverted_index.siblings, | 198 |
| abstract_inverted_index.software. | 103 |
| abstract_inverted_index.Additional | 0 |
| abstract_inverted_index.CIRCLE-seq | 95, 115 |
| abstract_inverted_index.CRISPRScan | 113, 129 |
| abstract_inverted_index.efficiency | 62 |
| abstract_inverted_index.frameshift | 154 |
| abstract_inverted_index.identified | 156 |
| abstract_inverted_index.percentage | 122 |
| abstract_inverted_index.prediction | 73 |
| abstract_inverted_index.reference. | 174 |
| abstract_inverted_index.sequencing | 22, 82, 99 |
| abstract_inverted_index.throughout | 55 |
| abstract_inverted_index.treatments | 242 |
| abstract_inverted_index.uninjected | 171, 196, 219 |
| abstract_inverted_index.Description | 107 |
| abstract_inverted_index.information | 145 |
| abstract_inverted_index.off-targets | 110, 138 |
| abstract_inverted_index.percentages | 180 |
| abstract_inverted_index.upregulated | 236 |
| abstract_inverted_index.efficiencies | 7 |
| abstract_inverted_index.intersecting | 125 |
| abstract_inverted_index.Cas9-injected | 215 |
| abstract_inverted_index.Supplementary | 3, 15, 27, 46, 58, 104, 148, 175, 206, 226 |
| abstract_inverted_index.catalytically | 190 |
| abstract_inverted_index.deconvolution | 85 |
| abstract_inverted_index.CrispRVariants | 102 |
| abstract_inverted_index.batch-siblings | 172, 220 |
| abstract_inverted_index.differentially | 35 |
| abstract_inverted_index.protein-coding | 158 |
| abstract_inverted_index.Oligonucleotides | 49 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 6 |
| citation_normalized_percentile |