Additional file 2 of Genome‑wide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencing Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.6084/m9.figshare.16893115
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.6084/m9.figshare.16893115
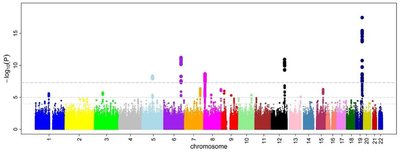
Additional file 2: Table S1. Information on the significant SNPs associated with 42 DW in the QTL region on chromosome 4. The data present the genomic location, effect, heritability, P value, and functional annotation of each GWAS significant SNP associated with 42 DW on chromosome 4. Table S2. Information on the significant SNPs associated with 84 DW in the QTL region on chromosome 4. The data present the genomic location, effect, heritability, P value, and functional annotation of each GWAS significant SNP associated with 84 DW on chromosome 4. Table S3. Information on the significant SNPs associated with ADG in the QTL region on chromosome 4. The data present the genomic location, effect, heritability, P value, and functional annotation of each GWAS significant SNP associated with ADG on chromosome 4. Table S4. Information on the significant SNPs associated with FCR in the QTL region on chromosome 4. The data present the genomic location, effect, heritability, P value, and functional annotation of each GWAS significant SNP associated with FCR on chromosome 4. Table S5. Average prediction inflation based on tenfold cross-validation. The data provide the average prediction inflation using ABLUP, GBLUP or GFBLUP models. Table S6. QTL contributions to the theoretical prediction accuracy (%). Description: The contributions were estimated by setting GWAS significant SNPs in each QTL as fixed factors. Table S7. Average prediction inflation addition after setting GWAS significant SNPs as fixed factors. The average prediction inflation changes after setting GWAS significant SNPs as fixed factors, where values approaching 0 represent lower inflations. Table S8. QTL contributions to heritability and observed prediction accuracy (%) using the GFBLUP model. The contributions were evaluated by estimating the contribution of two GRM using the GFBLUP model. Table S9. Average theoretical prediction accuracy using two GRM in the GFBLUP model. The data provide the average theoretical accuracy of prediction based on the GRM of GWAS significant sites and the remaining sites, respectively. Table S10. Average prediction inflation addition in the GFBLUP model. The changes in average prediction inflation evaluated by using the GRM without GWAS significant SNPs compared to that using both GRM, where values approaching 0 represent lower inflations.
Related Topics
- Type
- dataset
- Language
- en
- Landing Page
- https://doi.org/10.6084/m9.figshare.16893115
- OA Status
- gold
- References
- 72
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4394169230
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4394169230Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.6084/m9.figshare.16893115Digital Object Identifier
- Title
-
Additional file 2 of Genome‑wide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencingWork title
- Type
-
datasetOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-01-01Full publication date if available
- Authors
-
Ruifei Yang, Zhenqiang Xu, Qi Wang, Di Zhu, Cheng Bian, Jiangli Ren, Zhuolin Huang, Xiaoning Zhu, Zhixin Tian, Yuzhe Wang, Zi-Qin Jiang, Yiqiang Zhao, Dexiang Zhang, Ning Li, Xiaoxiang HuList of authors in order
- Landing page
-
https://doi.org/10.6084/m9.figshare.16893115Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.6084/m9.figshare.16893115Direct OA link when available
- Concepts
-
Plumage, Genotyping, Biology, Genome-wide association study, Genetics, Computational biology, Genome, Gene, Genotype, Single-nucleotide polymorphism, ZoologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
72Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4394169230 |
|---|---|
| doi | https://doi.org/10.6084/m9.figshare.16893115 |
| ids.doi | https://doi.org/10.6084/m9.figshare.16893115 |
| ids.openalex | https://openalex.org/W4394169230 |
| fwci | |
| type | dataset |
| title | Additional file 2 of Genome‑wide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencing |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10594 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9926000237464905 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic and phenotypic traits in livestock |
| topics[1].id | https://openalex.org/T10152 |
| topics[1].field.id | https://openalex.org/fields/11 |
| topics[1].field.display_name | Agricultural and Biological Sciences |
| topics[1].score | 0.9873999953269958 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1103 |
| topics[1].subfield.display_name | Animal Science and Zoology |
| topics[1].display_name | Animal Nutrition and Physiology |
| topics[2].id | https://openalex.org/T11468 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9775999784469604 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genetic Mapping and Diversity in Plants and Animals |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2779594111 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8506917953491211 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q3303665 |
| concepts[0].display_name | Plumage |
| concepts[1].id | https://openalex.org/C31467283 |
| concepts[1].level | 4 |
| concepts[1].score | 0.7701860666275024 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q912147 |
| concepts[1].display_name | Genotyping |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6930188536643982 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C106208931 |
| concepts[3].level | 5 |
| concepts[3].score | 0.5833289623260498 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[3].display_name | Genome-wide association study |
| concepts[4].id | https://openalex.org/C54355233 |
| concepts[4].level | 1 |
| concepts[4].score | 0.547236979007721 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[4].display_name | Genetics |
| concepts[5].id | https://openalex.org/C70721500 |
| concepts[5].level | 1 |
| concepts[5].score | 0.45588502287864685 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[5].display_name | Computational biology |
| concepts[6].id | https://openalex.org/C141231307 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4279704988002777 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[6].display_name | Genome |
| concepts[7].id | https://openalex.org/C104317684 |
| concepts[7].level | 2 |
| concepts[7].score | 0.26753413677215576 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[7].display_name | Gene |
| concepts[8].id | https://openalex.org/C135763542 |
| concepts[8].level | 3 |
| concepts[8].score | 0.24417340755462646 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[8].display_name | Genotype |
| concepts[9].id | https://openalex.org/C153209595 |
| concepts[9].level | 4 |
| concepts[9].score | 0.21327444911003113 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[9].display_name | Single-nucleotide polymorphism |
| concepts[10].id | https://openalex.org/C90856448 |
| concepts[10].level | 1 |
| concepts[10].score | 0.1390499472618103 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q431 |
| concepts[10].display_name | Zoology |
| keywords[0].id | https://openalex.org/keywords/plumage |
| keywords[0].score | 0.8506917953491211 |
| keywords[0].display_name | Plumage |
| keywords[1].id | https://openalex.org/keywords/genotyping |
| keywords[1].score | 0.7701860666275024 |
| keywords[1].display_name | Genotyping |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.6930188536643982 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[3].score | 0.5833289623260498 |
| keywords[3].display_name | Genome-wide association study |
| keywords[4].id | https://openalex.org/keywords/genetics |
| keywords[4].score | 0.547236979007721 |
| keywords[4].display_name | Genetics |
| keywords[5].id | https://openalex.org/keywords/computational-biology |
| keywords[5].score | 0.45588502287864685 |
| keywords[5].display_name | Computational biology |
| keywords[6].id | https://openalex.org/keywords/genome |
| keywords[6].score | 0.4279704988002777 |
| keywords[6].display_name | Genome |
| keywords[7].id | https://openalex.org/keywords/gene |
| keywords[7].score | 0.26753413677215576 |
| keywords[7].display_name | Gene |
| keywords[8].id | https://openalex.org/keywords/genotype |
| keywords[8].score | 0.24417340755462646 |
| keywords[8].display_name | Genotype |
| keywords[9].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[9].score | 0.21327444911003113 |
| keywords[9].display_name | Single-nucleotide polymorphism |
| keywords[10].id | https://openalex.org/keywords/zoology |
| keywords[10].score | 0.1390499472618103 |
| keywords[10].display_name | Zoology |
| language | en |
| locations[0].id | doi:10.6084/m9.figshare.16893115 |
| locations[0].is_oa | True |
| locations[0].source | |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | dataset |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.6084/m9.figshare.16893115 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5033570708 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-2074-6526 |
| authorships[0].author.display_name | Ruifei Yang |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Ruifei Yang |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5103248849 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-3328-9547 |
| authorships[1].author.display_name | Zhenqiang Xu |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Zhenqiang Xu |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5111103719 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-1058-5068 |
| authorships[2].author.display_name | Qi Wang |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Qi Wang |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5059413376 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-9362-3834 |
| authorships[3].author.display_name | Di Zhu |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Di Zhu |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5079061614 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-3638-5550 |
| authorships[4].author.display_name | Cheng Bian |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Cheng Bian |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5016969638 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Jiangli Ren |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Jiangli Ren |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5059576000 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Zhuolin Huang |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Zhuolin Huang |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5040869141 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-5743-879X |
| authorships[7].author.display_name | Xiaoning Zhu |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Xiaoning Zhu |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5042231897 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-2877-8282 |
| authorships[8].author.display_name | Zhixin Tian |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Zhixin Tian |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5100780554 |
| authorships[9].author.orcid | https://orcid.org/0000-0003-3474-7957 |
| authorships[9].author.display_name | Yuzhe Wang |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Yuzhe Wang |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5037383326 |
| authorships[10].author.orcid | |
| authorships[10].author.display_name | Zi-Qin Jiang |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Ziqin Jiang |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5001120955 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-0076-3476 |
| authorships[11].author.display_name | Yiqiang Zhao |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Yiqiang Zhao |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5069860925 |
| authorships[12].author.orcid | https://orcid.org/0009-0008-7144-3761 |
| authorships[12].author.display_name | Dexiang Zhang |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Dexiang Zhang |
| authorships[12].is_corresponding | False |
| authorships[13].author.id | https://openalex.org/A5053569900 |
| authorships[13].author.orcid | https://orcid.org/0000-0002-7721-8222 |
| authorships[13].author.display_name | Ning Li |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Ning Li |
| authorships[13].is_corresponding | False |
| authorships[14].author.id | https://openalex.org/A5020590657 |
| authorships[14].author.orcid | https://orcid.org/0000-0001-7045-0283 |
| authorships[14].author.display_name | Xiaoxiang Hu |
| authorships[14].author_position | last |
| authorships[14].raw_author_name | Xiaoxiang Hu |
| authorships[14].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.6084/m9.figshare.16893115 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Additional file 2 of Genome‑wide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencing |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T10594 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9926000237464905 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic and phenotypic traits in livestock |
| related_works | https://openalex.org/W1540487019, https://openalex.org/W2041851003, https://openalex.org/W2150709351, https://openalex.org/W2470640321, https://openalex.org/W4385415126, https://openalex.org/W2402073367, https://openalex.org/W2132123695, https://openalex.org/W2052461044, https://openalex.org/W4310191257, https://openalex.org/W1993755972 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.6084/m9.figshare.16893115 |
| best_oa_location.is_oa | True |
| best_oa_location.source | |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | dataset |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.6084/m9.figshare.16893115 |
| primary_location.id | doi:10.6084/m9.figshare.16893115 |
| primary_location.is_oa | True |
| primary_location.source | |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | dataset |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.6084/m9.figshare.16893115 |
| publication_date | 2021-01-01 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2060293383, https://openalex.org/W2094198395, https://openalex.org/W1962321979, https://openalex.org/W2738510634, https://openalex.org/W1928998639, https://openalex.org/W2108583185, https://openalex.org/W2023673366, https://openalex.org/W2334453152, https://openalex.org/W2110798717, https://openalex.org/W2767520269, https://openalex.org/W2095987942, https://openalex.org/W1825860353, https://openalex.org/W3019408181, https://openalex.org/W2899867539, https://openalex.org/W1972596659, https://openalex.org/W2491736610, https://openalex.org/W2807724764, https://openalex.org/W2623348960, https://openalex.org/W1972216668, https://openalex.org/W2952113787, https://openalex.org/W2033151532, https://openalex.org/W2161633633, https://openalex.org/W3042318275, https://openalex.org/W2038260948, https://openalex.org/W2254979106, https://openalex.org/W2401569012, https://openalex.org/W2142727510, https://openalex.org/W2765094284, https://openalex.org/W2600955197, https://openalex.org/W1956441033, https://openalex.org/W2924036988, https://openalex.org/W2020320027, https://openalex.org/W2580517199, https://openalex.org/W2886378237, https://openalex.org/W1839107343, https://openalex.org/W2184774706, https://openalex.org/W2028735580, https://openalex.org/W2952218813, https://openalex.org/W2801933709, https://openalex.org/W2968458064, https://openalex.org/W2096128561, https://openalex.org/W2606560121, https://openalex.org/W2377407741, https://openalex.org/W2912890533, https://openalex.org/W2190960123, https://openalex.org/W2156215519, https://openalex.org/W1581268762, https://openalex.org/W2063090075, https://openalex.org/W2533368445, https://openalex.org/W2810562347, https://openalex.org/W2981098318, https://openalex.org/W1965621126, https://openalex.org/W2991875949, https://openalex.org/W2626606138, https://openalex.org/W3081879876, https://openalex.org/W2316360588, https://openalex.org/W2583919829, https://openalex.org/W2160607966, https://openalex.org/W2077763475, https://openalex.org/W2885778610, https://openalex.org/W1645227518, https://openalex.org/W2077668635, https://openalex.org/W2035162791, https://openalex.org/W767574118, https://openalex.org/W2981346658, https://openalex.org/W2528815158, https://openalex.org/W2087761312, https://openalex.org/W1977950286, https://openalex.org/W2955612853, https://openalex.org/W3108090783, https://openalex.org/W2931834045, https://openalex.org/W2973029867 |
| referenced_works_count | 72 |
| abstract_inverted_index.0 | 249, 352 |
| abstract_inverted_index.P | 29, 72, 114, 155 |
| abstract_inverted_index.2: | 2 |
| abstract_inverted_index.4. | 20, 45, 63, 88, 105, 129, 146, 170 |
| abstract_inverted_index.42 | 12, 41 |
| abstract_inverted_index.84 | 55, 84 |
| abstract_inverted_index.DW | 13, 42, 56, 85 |
| abstract_inverted_index.as | 216, 230, 243 |
| abstract_inverted_index.by | 208, 272, 335 |
| abstract_inverted_index.in | 14, 57, 99, 140, 213, 292, 324, 330 |
| abstract_inverted_index.of | 34, 77, 119, 160, 276, 303, 309 |
| abstract_inverted_index.on | 6, 18, 43, 49, 61, 86, 92, 103, 127, 133, 144, 168, 177, 306 |
| abstract_inverted_index.or | 190 |
| abstract_inverted_index.to | 197, 257, 344 |
| abstract_inverted_index.(%) | 263 |
| abstract_inverted_index.ADG | 98, 126 |
| abstract_inverted_index.FCR | 139, 167 |
| abstract_inverted_index.GRM | 278, 291, 308, 338 |
| abstract_inverted_index.QTL | 16, 59, 101, 142, 195, 215, 255 |
| abstract_inverted_index.S1. | 4 |
| abstract_inverted_index.S2. | 47 |
| abstract_inverted_index.S3. | 90 |
| abstract_inverted_index.S4. | 131 |
| abstract_inverted_index.S5. | 172 |
| abstract_inverted_index.S6. | 194 |
| abstract_inverted_index.S7. | 220 |
| abstract_inverted_index.S8. | 254 |
| abstract_inverted_index.S9. | 284 |
| abstract_inverted_index.SNP | 38, 81, 123, 164 |
| abstract_inverted_index.The | 21, 64, 106, 147, 180, 204, 233, 268, 296, 328 |
| abstract_inverted_index.and | 31, 74, 116, 157, 259, 313 |
| abstract_inverted_index.the | 7, 15, 24, 50, 58, 67, 93, 100, 109, 134, 141, 150, 183, 198, 265, 274, 280, 293, 299, 307, 314, 325, 337 |
| abstract_inverted_index.two | 277, 290 |
| abstract_inverted_index.(%). | 202 |
| abstract_inverted_index.GRM, | 348 |
| abstract_inverted_index.GWAS | 36, 79, 121, 162, 210, 227, 240, 310, 340 |
| abstract_inverted_index.S10. | 319 |
| abstract_inverted_index.SNPs | 9, 52, 95, 136, 212, 229, 242, 342 |
| abstract_inverted_index.both | 347 |
| abstract_inverted_index.data | 22, 65, 107, 148, 181, 297 |
| abstract_inverted_index.each | 35, 78, 120, 161, 214 |
| abstract_inverted_index.file | 1 |
| abstract_inverted_index.that | 345 |
| abstract_inverted_index.were | 206, 270 |
| abstract_inverted_index.with | 11, 40, 54, 83, 97, 125, 138, 166 |
| abstract_inverted_index.GBLUP | 189 |
| abstract_inverted_index.Table | 3, 46, 89, 130, 171, 193, 219, 253, 283, 318 |
| abstract_inverted_index.after | 225, 238 |
| abstract_inverted_index.based | 176, 305 |
| abstract_inverted_index.fixed | 217, 231, 244 |
| abstract_inverted_index.lower | 251, 354 |
| abstract_inverted_index.sites | 312 |
| abstract_inverted_index.using | 187, 264, 279, 289, 336, 346 |
| abstract_inverted_index.where | 246, 349 |
| abstract_inverted_index.ABLUP, | 188 |
| abstract_inverted_index.GFBLUP | 191, 266, 281, 294, 326 |
| abstract_inverted_index.model. | 267, 282, 295, 327 |
| abstract_inverted_index.region | 17, 60, 102, 143 |
| abstract_inverted_index.sites, | 316 |
| abstract_inverted_index.value, | 30, 73, 115, 156 |
| abstract_inverted_index.values | 247, 350 |
| abstract_inverted_index.Average | 173, 221, 285, 320 |
| abstract_inverted_index.average | 184, 234, 300, 331 |
| abstract_inverted_index.changes | 237, 329 |
| abstract_inverted_index.effect, | 27, 70, 112, 153 |
| abstract_inverted_index.genomic | 25, 68, 110, 151 |
| abstract_inverted_index.models. | 192 |
| abstract_inverted_index.present | 23, 66, 108, 149 |
| abstract_inverted_index.provide | 182, 298 |
| abstract_inverted_index.setting | 209, 226, 239 |
| abstract_inverted_index.tenfold | 178 |
| abstract_inverted_index.without | 339 |
| abstract_inverted_index.accuracy | 201, 262, 288, 302 |
| abstract_inverted_index.addition | 224, 323 |
| abstract_inverted_index.compared | 343 |
| abstract_inverted_index.factors, | 245 |
| abstract_inverted_index.factors. | 218, 232 |
| abstract_inverted_index.observed | 260 |
| abstract_inverted_index.estimated | 207 |
| abstract_inverted_index.evaluated | 271, 334 |
| abstract_inverted_index.inflation | 175, 186, 223, 236, 322, 333 |
| abstract_inverted_index.location, | 26, 69, 111, 152 |
| abstract_inverted_index.remaining | 315 |
| abstract_inverted_index.represent | 250, 353 |
| abstract_inverted_index.Additional | 0 |
| abstract_inverted_index.annotation | 33, 76, 118, 159 |
| abstract_inverted_index.associated | 10, 39, 53, 82, 96, 124, 137, 165 |
| abstract_inverted_index.chromosome | 19, 44, 62, 87, 104, 128, 145, 169 |
| abstract_inverted_index.estimating | 273 |
| abstract_inverted_index.functional | 32, 75, 117, 158 |
| abstract_inverted_index.prediction | 174, 185, 200, 222, 235, 261, 287, 304, 321, 332 |
| abstract_inverted_index.Information | 5, 48, 91, 132 |
| abstract_inverted_index.approaching | 248, 351 |
| abstract_inverted_index.inflations. | 252, 355 |
| abstract_inverted_index.significant | 8, 37, 51, 80, 94, 122, 135, 163, 211, 228, 241, 311, 341 |
| abstract_inverted_index.theoretical | 199, 286, 301 |
| abstract_inverted_index.Description: | 203 |
| abstract_inverted_index.contribution | 275 |
| abstract_inverted_index.heritability | 258 |
| abstract_inverted_index.contributions | 196, 205, 256, 269 |
| abstract_inverted_index.heritability, | 28, 71, 113, 154 |
| abstract_inverted_index.respectively. | 317 |
| abstract_inverted_index.cross-validation. | 179 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 15 |
| citation_normalized_percentile |