All-optical voltage imaging-guided postsynaptic single-cell transcriptome profiling with Voltage-Seq Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.11.24.568588
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.11.24.568588
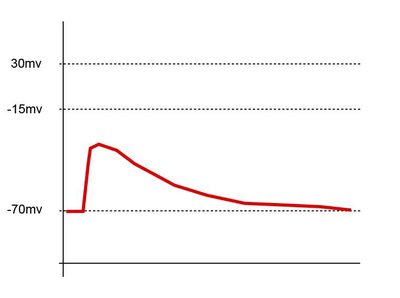
Neuronal pathways recruit large postsynaptic populations and maintain connections via distinct postsynaptic response types (PRTs). Until recently, PRTs were only accessible as a selection criterion for single-cell RNA-sequencing (scRNA-seq) through probing by low-throughput whole-cell electrophysiology. To overcome these limitations and target neurons based on specific PRTs for soma collection and subsequent scRNA-seq, we developed Voltage-Seq. An on-site analysis tool, VoltView, was created to guide soma harvesting of specific PRTs using a classifier based on a previously acquired connectome database from multiple animals. Here, we present a detailed step-by-step protocol, including setting up the optical path, the imaging setup, detailing the imaging procedure, and analysis, the reagents and protocols, complete procedure for sequencing library preparation, and provide other tips and pitfalls to assist researchers in conducting high-throughput all-optical voltage imaging, and to obtain the single-cell transcriptomic data from selected postsynaptic neurons. Voltage-seq workflow can be completed in ∼6 weeks including 4-5 weeks of viral expression of the Voltron sensor. The approach allows researchers to resolve the connectivity ratio of a specific pathway and explore the diversity of PRTs within that connectome. Furthermore, the high throughput in conjunction with quick analysis gives unique access to find specific connections within a large postsynaptic connectome. Voltage-seq also allows the investigation of correlations between connectivity and gene expression changes in a postsynaptic cell-type-specific manner in both excitatory and inhibitory connections.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.11.24.568588
- https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdf
- OA Status
- green
- References
- 12
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4389058809
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4389058809Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.11.24.568588Digital Object Identifier
- Title
-
All-optical voltage imaging-guided postsynaptic single-cell transcriptome profiling with Voltage-SeqWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-11-27Full publication date if available
- Authors
-
Veronika Csillag, J. C. Noble, Daniela Calvigioni, Björn Reinius, János FuzikList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.11.24.568588Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdfDirect OA link when available
- Concepts
-
Postsynaptic potential, Computer science, Connectome, Neuroscience, Soma, Excitatory postsynaptic potential, Inhibitory postsynaptic potential, Biology, Receptor, Functional connectivity, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
12Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4389058809 |
|---|---|
| doi | https://doi.org/10.1101/2023.11.24.568588 |
| ids.doi | https://doi.org/10.1101/2023.11.24.568588 |
| ids.openalex | https://openalex.org/W4389058809 |
| fwci | 0.0 |
| type | preprint |
| title | All-optical voltage imaging-guided postsynaptic single-cell transcriptome profiling with Voltage-Seq |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11289 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9984999895095825 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Single-cell and spatial transcriptomics |
| topics[1].id | https://openalex.org/T10581 |
| topics[1].field.id | https://openalex.org/fields/28 |
| topics[1].field.display_name | Neuroscience |
| topics[1].score | 0.9979000091552734 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2805 |
| topics[1].subfield.display_name | Cognitive Neuroscience |
| topics[1].display_name | Neural dynamics and brain function |
| topics[2].id | https://openalex.org/T10540 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9911999702453613 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1304 |
| topics[2].subfield.display_name | Biophysics |
| topics[2].display_name | Advanced Fluorescence Microscopy Techniques |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C197341189 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7240562438964844 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q863533 |
| concepts[0].display_name | Postsynaptic potential |
| concepts[1].id | https://openalex.org/C41008148 |
| concepts[1].level | 0 |
| concepts[1].score | 0.5471469759941101 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[1].display_name | Computer science |
| concepts[2].id | https://openalex.org/C45715564 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5049486756324768 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q1292103 |
| concepts[2].display_name | Connectome |
| concepts[3].id | https://openalex.org/C169760540 |
| concepts[3].level | 1 |
| concepts[3].score | 0.4733484089374542 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q207011 |
| concepts[3].display_name | Neuroscience |
| concepts[4].id | https://openalex.org/C2779617337 |
| concepts[4].level | 2 |
| concepts[4].score | 0.45818769931793213 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q842429 |
| concepts[4].display_name | Soma |
| concepts[5].id | https://openalex.org/C112592302 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4263647496700287 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1207387 |
| concepts[5].display_name | Excitatory postsynaptic potential |
| concepts[6].id | https://openalex.org/C17077164 |
| concepts[6].level | 2 |
| concepts[6].score | 0.32116347551345825 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1185869 |
| concepts[6].display_name | Inhibitory postsynaptic potential |
| concepts[7].id | https://openalex.org/C86803240 |
| concepts[7].level | 0 |
| concepts[7].score | 0.3004133701324463 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[7].display_name | Biology |
| concepts[8].id | https://openalex.org/C170493617 |
| concepts[8].level | 2 |
| concepts[8].score | 0.0 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[8].display_name | Receptor |
| concepts[9].id | https://openalex.org/C3018011982 |
| concepts[9].level | 2 |
| concepts[9].score | 0.0 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7316120 |
| concepts[9].display_name | Functional connectivity |
| concepts[10].id | https://openalex.org/C55493867 |
| concepts[10].level | 1 |
| concepts[10].score | 0.0 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[10].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/postsynaptic-potential |
| keywords[0].score | 0.7240562438964844 |
| keywords[0].display_name | Postsynaptic potential |
| keywords[1].id | https://openalex.org/keywords/computer-science |
| keywords[1].score | 0.5471469759941101 |
| keywords[1].display_name | Computer science |
| keywords[2].id | https://openalex.org/keywords/connectome |
| keywords[2].score | 0.5049486756324768 |
| keywords[2].display_name | Connectome |
| keywords[3].id | https://openalex.org/keywords/neuroscience |
| keywords[3].score | 0.4733484089374542 |
| keywords[3].display_name | Neuroscience |
| keywords[4].id | https://openalex.org/keywords/soma |
| keywords[4].score | 0.45818769931793213 |
| keywords[4].display_name | Soma |
| keywords[5].id | https://openalex.org/keywords/excitatory-postsynaptic-potential |
| keywords[5].score | 0.4263647496700287 |
| keywords[5].display_name | Excitatory postsynaptic potential |
| keywords[6].id | https://openalex.org/keywords/inhibitory-postsynaptic-potential |
| keywords[6].score | 0.32116347551345825 |
| keywords[6].display_name | Inhibitory postsynaptic potential |
| keywords[7].id | https://openalex.org/keywords/biology |
| keywords[7].score | 0.3004133701324463 |
| keywords[7].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1101/2023.11.24.568588 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.11.24.568588 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5071892658 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Veronika Csillag |
| authorships[0].countries | SE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden |
| authorships[0].institutions[0].id | https://openalex.org/I28166907 |
| authorships[0].institutions[0].ror | https://ror.org/056d84691 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[0].institutions[0].country_code | SE |
| authorships[0].institutions[0].display_name | Karolinska Institutet |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Veronika Csillag |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden |
| authorships[1].author.id | https://openalex.org/A5012396032 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-4450-429X |
| authorships[1].author.display_name | J. C. Noble |
| authorships[1].countries | SE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden |
| authorships[1].institutions[0].id | https://openalex.org/I28166907 |
| authorships[1].institutions[0].ror | https://ror.org/056d84691 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[1].institutions[0].country_code | SE |
| authorships[1].institutions[0].display_name | Karolinska Institutet |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | JC Noble |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden |
| authorships[2].author.id | https://openalex.org/A5075549429 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-6122-9744 |
| authorships[2].author.display_name | Daniela Calvigioni |
| authorships[2].countries | SE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden |
| authorships[2].institutions[0].id | https://openalex.org/I28166907 |
| authorships[2].institutions[0].ror | https://ror.org/056d84691 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[2].institutions[0].country_code | SE |
| authorships[2].institutions[0].display_name | Karolinska Institutet |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Daniela Calvigioni |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden |
| authorships[3].author.id | https://openalex.org/A5052918715 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-7021-5248 |
| authorships[3].author.display_name | Björn Reinius |
| authorships[3].countries | SE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden |
| authorships[3].institutions[0].id | https://openalex.org/I28166907 |
| authorships[3].institutions[0].ror | https://ror.org/056d84691 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[3].institutions[0].country_code | SE |
| authorships[3].institutions[0].display_name | Karolinska Institutet |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Björn Reinius |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden |
| authorships[4].author.id | https://openalex.org/A5056549934 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-5408-4882 |
| authorships[4].author.display_name | János Fuzik |
| authorships[4].countries | SE |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden |
| authorships[4].institutions[0].id | https://openalex.org/I28166907 |
| authorships[4].institutions[0].ror | https://ror.org/056d84691 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[4].institutions[0].country_code | SE |
| authorships[4].institutions[0].display_name | Karolinska Institutet |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | János Fuzik |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | All-optical voltage imaging-guided postsynaptic single-cell transcriptome profiling with Voltage-Seq |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11289 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9984999895095825 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Single-cell and spatial transcriptomics |
| related_works | https://openalex.org/W2950596497, https://openalex.org/W2436760910, https://openalex.org/W4303432678, https://openalex.org/W2043066229, https://openalex.org/W2142526561, https://openalex.org/W2974163011, https://openalex.org/W2023878078, https://openalex.org/W4285127779, https://openalex.org/W2082329602, https://openalex.org/W1546859207 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2023.11.24.568588 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.11.24.568588 |
| primary_location.id | doi:10.1101/2023.11.24.568588 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/11/27/2023.11.24.568588.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.11.24.568588 |
| publication_date | 2023-11-27 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2268971953, https://openalex.org/W2267554996, https://openalex.org/W4303699451, https://openalex.org/W2964363746, https://openalex.org/W2908737449, https://openalex.org/W1982277188, https://openalex.org/W3111521801, https://openalex.org/W4384925173, https://openalex.org/W2788172302, https://openalex.org/W2019552331, https://openalex.org/W1526404082, https://openalex.org/W2026863186 |
| referenced_works_count | 12 |
| abstract_inverted_index.a | 23, 71, 75, 86, 169, 198, 216 |
| abstract_inverted_index.An | 56 |
| abstract_inverted_index.To | 36 |
| abstract_inverted_index.as | 22 |
| abstract_inverted_index.be | 144 |
| abstract_inverted_index.by | 32 |
| abstract_inverted_index.in | 124, 146, 185, 215, 220 |
| abstract_inverted_index.of | 67, 152, 155, 168, 176, 207 |
| abstract_inverted_index.on | 44, 74 |
| abstract_inverted_index.to | 63, 121, 131, 163, 193 |
| abstract_inverted_index.up | 92 |
| abstract_inverted_index.we | 53, 84 |
| abstract_inverted_index.4-5 | 150 |
| abstract_inverted_index.The | 159 |
| abstract_inverted_index.and | 7, 40, 50, 103, 107, 115, 119, 130, 172, 211, 223 |
| abstract_inverted_index.can | 143 |
| abstract_inverted_index.for | 26, 47, 111 |
| abstract_inverted_index.the | 93, 96, 100, 105, 133, 156, 165, 174, 182, 205 |
| abstract_inverted_index.via | 10 |
| abstract_inverted_index.was | 61 |
| abstract_inverted_index.PRTs | 18, 46, 69, 177 |
| abstract_inverted_index.also | 203 |
| abstract_inverted_index.both | 221 |
| abstract_inverted_index.data | 136 |
| abstract_inverted_index.find | 194 |
| abstract_inverted_index.from | 80, 137 |
| abstract_inverted_index.gene | 212 |
| abstract_inverted_index.high | 183 |
| abstract_inverted_index.only | 20 |
| abstract_inverted_index.soma | 48, 65 |
| abstract_inverted_index.that | 179 |
| abstract_inverted_index.tips | 118 |
| abstract_inverted_index.were | 19 |
| abstract_inverted_index.with | 187 |
| abstract_inverted_index.∼6 | 147 |
| abstract_inverted_index.Here, | 83 |
| abstract_inverted_index.Until | 16 |
| abstract_inverted_index.based | 43, 73 |
| abstract_inverted_index.gives | 190 |
| abstract_inverted_index.guide | 64 |
| abstract_inverted_index.large | 4, 199 |
| abstract_inverted_index.other | 117 |
| abstract_inverted_index.path, | 95 |
| abstract_inverted_index.quick | 188 |
| abstract_inverted_index.ratio | 167 |
| abstract_inverted_index.these | 38 |
| abstract_inverted_index.tool, | 59 |
| abstract_inverted_index.types | 14 |
| abstract_inverted_index.using | 70 |
| abstract_inverted_index.viral | 153 |
| abstract_inverted_index.weeks | 148, 151 |
| abstract_inverted_index.access | 192 |
| abstract_inverted_index.allows | 161, 204 |
| abstract_inverted_index.assist | 122 |
| abstract_inverted_index.manner | 219 |
| abstract_inverted_index.obtain | 132 |
| abstract_inverted_index.setup, | 98 |
| abstract_inverted_index.target | 41 |
| abstract_inverted_index.unique | 191 |
| abstract_inverted_index.within | 178, 197 |
| abstract_inverted_index.(PRTs). | 15 |
| abstract_inverted_index.Voltron | 157 |
| abstract_inverted_index.between | 209 |
| abstract_inverted_index.changes | 214 |
| abstract_inverted_index.created | 62 |
| abstract_inverted_index.explore | 173 |
| abstract_inverted_index.imaging | 97, 101 |
| abstract_inverted_index.library | 113 |
| abstract_inverted_index.neurons | 42 |
| abstract_inverted_index.on-site | 57 |
| abstract_inverted_index.optical | 94 |
| abstract_inverted_index.pathway | 171 |
| abstract_inverted_index.present | 85 |
| abstract_inverted_index.probing | 31 |
| abstract_inverted_index.provide | 116 |
| abstract_inverted_index.recruit | 3 |
| abstract_inverted_index.resolve | 164 |
| abstract_inverted_index.sensor. | 158 |
| abstract_inverted_index.setting | 91 |
| abstract_inverted_index.through | 30 |
| abstract_inverted_index.voltage | 128 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Neuronal | 1 |
| abstract_inverted_index.acquired | 77 |
| abstract_inverted_index.analysis | 58, 189 |
| abstract_inverted_index.animals. | 82 |
| abstract_inverted_index.approach | 160 |
| abstract_inverted_index.complete | 109 |
| abstract_inverted_index.database | 79 |
| abstract_inverted_index.detailed | 87 |
| abstract_inverted_index.distinct | 11 |
| abstract_inverted_index.imaging, | 129 |
| abstract_inverted_index.maintain | 8 |
| abstract_inverted_index.multiple | 81 |
| abstract_inverted_index.neurons. | 140 |
| abstract_inverted_index.overcome | 37 |
| abstract_inverted_index.pathways | 2 |
| abstract_inverted_index.pitfalls | 120 |
| abstract_inverted_index.reagents | 106 |
| abstract_inverted_index.response | 13 |
| abstract_inverted_index.selected | 138 |
| abstract_inverted_index.specific | 45, 68, 170, 195 |
| abstract_inverted_index.workflow | 142 |
| abstract_inverted_index.VoltView, | 60 |
| abstract_inverted_index.analysis, | 104 |
| abstract_inverted_index.completed | 145 |
| abstract_inverted_index.criterion | 25 |
| abstract_inverted_index.detailing | 99 |
| abstract_inverted_index.developed | 54 |
| abstract_inverted_index.diversity | 175 |
| abstract_inverted_index.including | 90, 149 |
| abstract_inverted_index.procedure | 110 |
| abstract_inverted_index.protocol, | 89 |
| abstract_inverted_index.recently, | 17 |
| abstract_inverted_index.selection | 24 |
| abstract_inverted_index.accessible | 21 |
| abstract_inverted_index.classifier | 72 |
| abstract_inverted_index.collection | 49 |
| abstract_inverted_index.conducting | 125 |
| abstract_inverted_index.connectome | 78 |
| abstract_inverted_index.excitatory | 222 |
| abstract_inverted_index.expression | 154, 213 |
| abstract_inverted_index.harvesting | 66 |
| abstract_inverted_index.inhibitory | 224 |
| abstract_inverted_index.previously | 76 |
| abstract_inverted_index.procedure, | 102 |
| abstract_inverted_index.protocols, | 108 |
| abstract_inverted_index.scRNA-seq, | 52 |
| abstract_inverted_index.sequencing | 112 |
| abstract_inverted_index.subsequent | 51 |
| abstract_inverted_index.throughput | 184 |
| abstract_inverted_index.whole-cell | 34 |
| abstract_inverted_index.(scRNA-seq) | 29 |
| abstract_inverted_index.Voltage-seq | 141, 202 |
| abstract_inverted_index.all-optical | 127 |
| abstract_inverted_index.conjunction | 186 |
| abstract_inverted_index.connections | 9, 196 |
| abstract_inverted_index.connectome. | 180, 201 |
| abstract_inverted_index.limitations | 39 |
| abstract_inverted_index.populations | 6 |
| abstract_inverted_index.researchers | 123, 162 |
| abstract_inverted_index.single-cell | 27, 134 |
| abstract_inverted_index.Furthermore, | 181 |
| abstract_inverted_index.Voltage-Seq. | 55 |
| abstract_inverted_index.connections. | 225 |
| abstract_inverted_index.connectivity | 166, 210 |
| abstract_inverted_index.correlations | 208 |
| abstract_inverted_index.postsynaptic | 5, 12, 139, 200, 217 |
| abstract_inverted_index.preparation, | 114 |
| abstract_inverted_index.step-by-step | 88 |
| abstract_inverted_index.investigation | 206 |
| abstract_inverted_index.RNA-sequencing | 28 |
| abstract_inverted_index.low-throughput | 33 |
| abstract_inverted_index.transcriptomic | 135 |
| abstract_inverted_index.high-throughput | 126 |
| abstract_inverted_index.cell-type-specific | 218 |
| abstract_inverted_index.electrophysiology. | 35 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5056549934 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| corresponding_institution_ids | https://openalex.org/I28166907 |
| citation_normalized_percentile.value | 0.33183713 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |