Amplification of Telomeric Arrays via Rolling-circle Mechanism Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.17615/k88m-6a49
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.17615/k88m-6a49
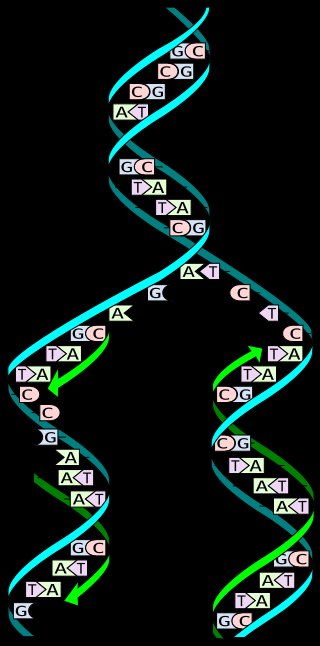
Alternative (telomerase-independent) lengthening of telomeres mediated through homologous recombination is often accompanied by a generation of extrachromosomal telomeric circles (t-circles), whose role in direct promotion of recombinational telomere elongation has been recently demonstrated. Here we present evidence that t-circles in a natural telomerase-deficient system of mitochondria of the yeast Candida parapsilosis replicate independently of the linear chromosome via a rolling-circle mechanism. This is supported by an observation of (i) single-stranded DNA consisting of concatameric arrays of telomeric sequence, (ii) lasso-shaped molecules representing rolling-circle intermediates, and (iii) preferential incorporation of deoxyribonucleotides into telomeric fragments and t-circles. Analysis of naturally occurring variant t-circles revealed conserved motifs with potential function in driving the rolling-circle replication. These data indicate that extrachromosomal t-circles observed in a wide variety of organisms, including yeasts, plants, Xenopus laevis, and certain human cell lines, may represent independent replicons generating telomeric sequences and, thus, actively participating in telomere dynamics. Moreover, because of the promiscuous occurrence of t-circles across phyla, the results from yeast mitochondria have implications related to the primordial system of telomere maintenance, providing a paradigm for evolution of telomeres in nuclei of early eukaryotes.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.17615/k88m-6a49
- OA Status
- green
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4301763323
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4301763323Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.17615/k88m-6a49Digital Object Identifier
- Title
-
Amplification of Telomeric Arrays via Rolling-circle MechanismWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-07-03Full publication date if available
- Authors
-
Jozef Nosek, Adriana Ryčovská, Alexander M. Makhov, Jack D. Griffith, Ľubomír TomáškaList of authors in order
- Landing page
-
https://doi.org/10.17615/k88m-6a49Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.17615/k88m-6a49Direct OA link when available
- Concepts
-
Rolling circle replication, Mechanism (biology), Materials science, Computer science, Biology, Genetics, Physics, DNA, DNA replication, Quantum mechanicsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4301763323 |
|---|---|
| doi | https://doi.org/10.17615/k88m-6a49 |
| ids.doi | https://doi.org/10.17615/k88m-6a49 |
| ids.openalex | https://openalex.org/W4301763323 |
| fwci | 0.0 |
| type | article |
| title | Amplification of Telomeric Arrays via Rolling-circle Mechanism |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11946 |
| topics[0].field.id | https://openalex.org/fields/22 |
| topics[0].field.display_name | Engineering |
| topics[0].score | 0.9977999925613403 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2202 |
| topics[0].subfield.display_name | Aerospace Engineering |
| topics[0].display_name | Antenna Design and Optimization |
| topics[1].id | https://openalex.org/T12162 |
| topics[1].field.id | https://openalex.org/fields/17 |
| topics[1].field.display_name | Computer Science |
| topics[1].score | 0.9764999747276306 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1703 |
| topics[1].subfield.display_name | Computational Theory and Mathematics |
| topics[1].display_name | Cellular Automata and Applications |
| topics[2].id | https://openalex.org/T11870 |
| topics[2].field.id | https://openalex.org/fields/22 |
| topics[2].field.display_name | Engineering |
| topics[2].score | 0.9459999799728394 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2205 |
| topics[2].subfield.display_name | Civil and Structural Engineering |
| topics[2].display_name | Structural Analysis and Optimization |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C7452308 |
| concepts[0].level | 4 |
| concepts[0].score | 0.7153230309486389 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q3112822 |
| concepts[0].display_name | Rolling circle replication |
| concepts[1].id | https://openalex.org/C89611455 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6610246896743774 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q6804646 |
| concepts[1].display_name | Mechanism (biology) |
| concepts[2].id | https://openalex.org/C192562407 |
| concepts[2].level | 0 |
| concepts[2].score | 0.3641089200973511 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q228736 |
| concepts[2].display_name | Materials science |
| concepts[3].id | https://openalex.org/C41008148 |
| concepts[3].level | 0 |
| concepts[3].score | 0.32245054841041565 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[3].display_name | Computer science |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.25461339950561523 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C54355233 |
| concepts[5].level | 1 |
| concepts[5].score | 0.2516809403896332 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[5].display_name | Genetics |
| concepts[6].id | https://openalex.org/C121332964 |
| concepts[6].level | 0 |
| concepts[6].score | 0.20675918459892273 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[6].display_name | Physics |
| concepts[7].id | https://openalex.org/C552990157 |
| concepts[7].level | 2 |
| concepts[7].score | 0.12434855103492737 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[7].display_name | DNA |
| concepts[8].id | https://openalex.org/C73573662 |
| concepts[8].level | 3 |
| concepts[8].score | 0.0 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q130996 |
| concepts[8].display_name | DNA replication |
| concepts[9].id | https://openalex.org/C62520636 |
| concepts[9].level | 1 |
| concepts[9].score | 0.0 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q944 |
| concepts[9].display_name | Quantum mechanics |
| keywords[0].id | https://openalex.org/keywords/rolling-circle-replication |
| keywords[0].score | 0.7153230309486389 |
| keywords[0].display_name | Rolling circle replication |
| keywords[1].id | https://openalex.org/keywords/mechanism |
| keywords[1].score | 0.6610246896743774 |
| keywords[1].display_name | Mechanism (biology) |
| keywords[2].id | https://openalex.org/keywords/materials-science |
| keywords[2].score | 0.3641089200973511 |
| keywords[2].display_name | Materials science |
| keywords[3].id | https://openalex.org/keywords/computer-science |
| keywords[3].score | 0.32245054841041565 |
| keywords[3].display_name | Computer science |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.25461339950561523 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/genetics |
| keywords[5].score | 0.2516809403896332 |
| keywords[5].display_name | Genetics |
| keywords[6].id | https://openalex.org/keywords/physics |
| keywords[6].score | 0.20675918459892273 |
| keywords[6].display_name | Physics |
| keywords[7].id | https://openalex.org/keywords/dna |
| keywords[7].score | 0.12434855103492737 |
| keywords[7].display_name | DNA |
| language | en |
| locations[0].id | doi:10.17615/k88m-6a49 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S7407051488 |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | UNC Libraries |
| locations[0].source.host_organization | |
| locations[0].source.host_organization_name | |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | article-journal |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.17615/k88m-6a49 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5041903432 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1020-5451 |
| authorships[0].author.display_name | Jozef Nosek |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Nosek, Jozef |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5011578361 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Adriana Ryčovská |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Rycovska, Adriana |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5026749590 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-7910-5766 |
| authorships[2].author.display_name | Alexander M. Makhov |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I114027177 |
| authorships[2].affiliations[0].raw_affiliation_string | University of North Carolina at Chapel Hill |
| authorships[2].institutions[0].id | https://openalex.org/I114027177 |
| authorships[2].institutions[0].ror | https://ror.org/0130frc33 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I114027177 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of North Carolina at Chapel Hill |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Makhov, Alexander M. |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | University of North Carolina at Chapel Hill |
| authorships[3].author.id | https://openalex.org/A5061660001 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-8357-8689 |
| authorships[3].author.display_name | Jack D. Griffith |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I114027177 |
| authorships[3].affiliations[0].raw_affiliation_string | University of North Carolina at Chapel Hill |
| authorships[3].institutions[0].id | https://openalex.org/I114027177 |
| authorships[3].institutions[0].ror | https://ror.org/0130frc33 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I114027177 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of North Carolina at Chapel Hill |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Griffith, Jack D. |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | University of North Carolina at Chapel Hill |
| authorships[4].author.id | https://openalex.org/A5012452039 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-4886-1910 |
| authorships[4].author.display_name | Ľubomír Tomáška |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I114027177 |
| authorships[4].affiliations[0].raw_affiliation_string | University of North Carolina at Chapel Hill |
| authorships[4].institutions[0].id | https://openalex.org/I114027177 |
| authorships[4].institutions[0].ror | https://ror.org/0130frc33 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I114027177 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | University of North Carolina at Chapel Hill |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Tomaska, Lubomir |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | University of North Carolina at Chapel Hill |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.17615/k88m-6a49 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2022-10-05T00:00:00 |
| display_name | Amplification of Telomeric Arrays via Rolling-circle Mechanism |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T11946 |
| primary_topic.field.id | https://openalex.org/fields/22 |
| primary_topic.field.display_name | Engineering |
| primary_topic.score | 0.9977999925613403 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2202 |
| primary_topic.subfield.display_name | Aerospace Engineering |
| primary_topic.display_name | Antenna Design and Optimization |
| related_works | https://openalex.org/W2899084033, https://openalex.org/W2748952813, https://openalex.org/W2179063564, https://openalex.org/W2171758033, https://openalex.org/W2996077980, https://openalex.org/W4212879511, https://openalex.org/W2737498735, https://openalex.org/W4311158810, https://openalex.org/W2160608618, https://openalex.org/W1987137857 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.17615/k88m-6a49 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S7407051488 |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | UNC Libraries |
| best_oa_location.source.host_organization | |
| best_oa_location.source.host_organization_name | |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | article-journal |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.17615/k88m-6a49 |
| primary_location.id | doi:10.17615/k88m-6a49 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S7407051488 |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | UNC Libraries |
| primary_location.source.host_organization | |
| primary_location.source.host_organization_name | |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | article-journal |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.17615/k88m-6a49 |
| publication_date | 2021-07-03 |
| publication_year | 2021 |
| referenced_works_count | 0 |
| abstract_inverted_index.a | 13, 40, 58, 120, 175 |
| abstract_inverted_index.an | 65 |
| abstract_inverted_index.by | 12, 64 |
| abstract_inverted_index.in | 22, 39, 107, 119, 146, 181 |
| abstract_inverted_index.is | 9, 62 |
| abstract_inverted_index.of | 3, 15, 25, 44, 46, 53, 67, 72, 75, 88, 96, 123, 151, 155, 171, 179, 183 |
| abstract_inverted_index.to | 167 |
| abstract_inverted_index.we | 34 |
| abstract_inverted_index.(i) | 68 |
| abstract_inverted_index.DNA | 70 |
| abstract_inverted_index.and | 84, 93, 130 |
| abstract_inverted_index.for | 177 |
| abstract_inverted_index.has | 29 |
| abstract_inverted_index.may | 135 |
| abstract_inverted_index.the | 47, 54, 109, 152, 159, 168 |
| abstract_inverted_index.via | 57 |
| abstract_inverted_index.(ii) | 78 |
| abstract_inverted_index.Here | 33 |
| abstract_inverted_index.This | 61 |
| abstract_inverted_index.and, | 142 |
| abstract_inverted_index.been | 30 |
| abstract_inverted_index.cell | 133 |
| abstract_inverted_index.data | 113 |
| abstract_inverted_index.from | 161 |
| abstract_inverted_index.have | 164 |
| abstract_inverted_index.into | 90 |
| abstract_inverted_index.role | 21 |
| abstract_inverted_index.that | 37, 115 |
| abstract_inverted_index.wide | 121 |
| abstract_inverted_index.with | 104 |
| abstract_inverted_index.(iii) | 85 |
| abstract_inverted_index.These | 112 |
| abstract_inverted_index.early | 184 |
| abstract_inverted_index.human | 132 |
| abstract_inverted_index.often | 10 |
| abstract_inverted_index.thus, | 143 |
| abstract_inverted_index.whose | 20 |
| abstract_inverted_index.yeast | 48, 162 |
| abstract_inverted_index.across | 157 |
| abstract_inverted_index.arrays | 74 |
| abstract_inverted_index.direct | 23 |
| abstract_inverted_index.linear | 55 |
| abstract_inverted_index.lines, | 134 |
| abstract_inverted_index.motifs | 103 |
| abstract_inverted_index.nuclei | 182 |
| abstract_inverted_index.phyla, | 158 |
| abstract_inverted_index.system | 43, 170 |
| abstract_inverted_index.Candida | 49 |
| abstract_inverted_index.Xenopus | 128 |
| abstract_inverted_index.because | 150 |
| abstract_inverted_index.certain | 131 |
| abstract_inverted_index.circles | 18 |
| abstract_inverted_index.driving | 108 |
| abstract_inverted_index.laevis, | 129 |
| abstract_inverted_index.natural | 41 |
| abstract_inverted_index.plants, | 127 |
| abstract_inverted_index.present | 35 |
| abstract_inverted_index.related | 166 |
| abstract_inverted_index.results | 160 |
| abstract_inverted_index.through | 6 |
| abstract_inverted_index.variant | 99 |
| abstract_inverted_index.variety | 122 |
| abstract_inverted_index.yeasts, | 126 |
| abstract_inverted_index.Analysis | 95 |
| abstract_inverted_index.actively | 144 |
| abstract_inverted_index.evidence | 36 |
| abstract_inverted_index.function | 106 |
| abstract_inverted_index.indicate | 114 |
| abstract_inverted_index.mediated | 5 |
| abstract_inverted_index.observed | 118 |
| abstract_inverted_index.paradigm | 176 |
| abstract_inverted_index.recently | 31 |
| abstract_inverted_index.revealed | 101 |
| abstract_inverted_index.telomere | 27, 147, 172 |
| abstract_inverted_index.Moreover, | 149 |
| abstract_inverted_index.conserved | 102 |
| abstract_inverted_index.dynamics. | 148 |
| abstract_inverted_index.evolution | 178 |
| abstract_inverted_index.fragments | 92 |
| abstract_inverted_index.including | 125 |
| abstract_inverted_index.molecules | 80 |
| abstract_inverted_index.naturally | 97 |
| abstract_inverted_index.occurring | 98 |
| abstract_inverted_index.potential | 105 |
| abstract_inverted_index.promotion | 24 |
| abstract_inverted_index.providing | 174 |
| abstract_inverted_index.replicate | 51 |
| abstract_inverted_index.replicons | 138 |
| abstract_inverted_index.represent | 136 |
| abstract_inverted_index.sequence, | 77 |
| abstract_inverted_index.sequences | 141 |
| abstract_inverted_index.supported | 63 |
| abstract_inverted_index.t-circles | 38, 100, 117, 156 |
| abstract_inverted_index.telomeres | 4, 180 |
| abstract_inverted_index.telomeric | 17, 76, 91, 140 |
| abstract_inverted_index.chromosome | 56 |
| abstract_inverted_index.consisting | 71 |
| abstract_inverted_index.elongation | 28 |
| abstract_inverted_index.generating | 139 |
| abstract_inverted_index.generation | 14 |
| abstract_inverted_index.homologous | 7 |
| abstract_inverted_index.mechanism. | 60 |
| abstract_inverted_index.occurrence | 154 |
| abstract_inverted_index.organisms, | 124 |
| abstract_inverted_index.primordial | 169 |
| abstract_inverted_index.t-circles. | 94 |
| abstract_inverted_index.Alternative | 0 |
| abstract_inverted_index.accompanied | 11 |
| abstract_inverted_index.eukaryotes. | 185 |
| abstract_inverted_index.independent | 137 |
| abstract_inverted_index.lengthening | 2 |
| abstract_inverted_index.observation | 66 |
| abstract_inverted_index.promiscuous | 153 |
| abstract_inverted_index.(t-circles), | 19 |
| abstract_inverted_index.concatameric | 73 |
| abstract_inverted_index.implications | 165 |
| abstract_inverted_index.lasso-shaped | 79 |
| abstract_inverted_index.maintenance, | 173 |
| abstract_inverted_index.mitochondria | 45, 163 |
| abstract_inverted_index.parapsilosis | 50 |
| abstract_inverted_index.preferential | 86 |
| abstract_inverted_index.replication. | 111 |
| abstract_inverted_index.representing | 81 |
| abstract_inverted_index.demonstrated. | 32 |
| abstract_inverted_index.incorporation | 87 |
| abstract_inverted_index.independently | 52 |
| abstract_inverted_index.participating | 145 |
| abstract_inverted_index.recombination | 8 |
| abstract_inverted_index.intermediates, | 83 |
| abstract_inverted_index.rolling-circle | 59, 82, 110 |
| abstract_inverted_index.recombinational | 26 |
| abstract_inverted_index.single-stranded | 69 |
| abstract_inverted_index.extrachromosomal | 16, 116 |
| abstract_inverted_index.deoxyribonucleotides | 89 |
| abstract_inverted_index.telomerase-deficient | 42 |
| abstract_inverted_index.(telomerase-independent) | 1 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| citation_normalized_percentile.value | 0.12265611 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |