Anticancer Target Combinations: Network-Informed Signaling-Based Approach to Discovery Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.10.11.617918
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.10.11.617918
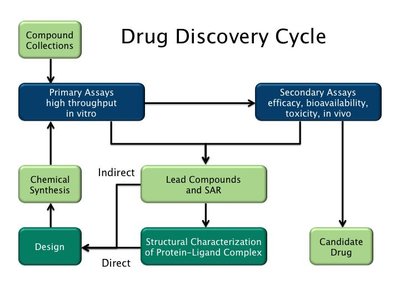
While anticancer drug discovery has seen dramatic innovations and successes, sequential single therapies are time-limited by resistance, and combinatorial strategies have been lagging. The number of possible drug combinations is vast. To select drug combinations the oncologist requires knowledge of the optimal combination of proteins to co-target. Currently, combinations that the oncologist considers are primarily from empirical observations and clinical praxis. Our aim is to develop a signaling-based method to discover optimal proteins for the oncologist to co-target with drug combinations, and test it on available, patient-derived data. To temper the expected resistance to single drug regimen, we offer a concept-based stratified pipeline aimed at selecting co-targets for drug combinations. Our strategy is unique in its co-target selection being based on signaling pathways. This is significant since in cancer, drug resistance commonly bypasses blocked proteins by wielding alternative, or complementary, routes to execute cell proliferation. Our network-informed signaling-based approach harnesses advanced network concepts and metrics, and our compiled, tissue-specific co-existing mutations. Co-existing driver mutations are common in resistance. Thus, to mimic cancer and counter drug resistance scenarios, our pipeline seeks co-targets that when targeted by drug combinations, can shut off cancer’s modus operandi . That is, its parallel or complementary signaling pathways would be blocked. Rotating through combinations could further lessen emerging resistance. We applied it to patient-derived breast and colorectal ESR1 | PIK3CA and BRAF | PIK3CA subnetworks. Consistently, in breast cancer, our results suggest co-targeting proteins from the ESR1|PIK3CA subnetwork with an alpelisib-LJM716 combination. In colorectal cancer, they co-target BRAF | PIK3CA with alpelisib, cetuximab, and encorafenib combination. Collectively, our pipeline’s results are promising, and validated by patient-based xenografts. GRAPHICAL ABSTRACT
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2024.10.11.617918
- https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdf
- OA Status
- green
- References
- 143
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4403421792
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4403421792Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2024.10.11.617918Digital Object Identifier
- Title
-
Anticancer Target Combinations: Network-Informed Signaling-Based Approach to DiscoveryWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-10-15Full publication date if available
- Authors
-
Bengi Ruken Yavuz, Hyunbum Jang, Ruth NussinovList of authors in order
- Landing page
-
https://doi.org/10.1101/2024.10.11.617918Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdfDirect OA link when available
- Concepts
-
Drug discovery, Computational biology, Computer science, Biology, BioinformaticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
143Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4403421792 |
|---|---|
| doi | https://doi.org/10.1101/2024.10.11.617918 |
| ids.doi | https://doi.org/10.1101/2024.10.11.617918 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/41306515 |
| ids.openalex | https://openalex.org/W4403421792 |
| fwci | 0.0 |
| type | preprint |
| title | Anticancer Target Combinations: Network-Informed Signaling-Based Approach to Discovery |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9980999827384949 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T10887 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9887999892234802 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Bioinformatics and Genomic Networks |
| topics[2].id | https://openalex.org/T12023 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9262999892234802 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2736 |
| topics[2].subfield.display_name | Pharmacology |
| topics[2].display_name | Cholinesterase and Neurodegenerative Diseases |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C74187038 |
| concepts[0].level | 2 |
| concepts[0].score | 0.41158172488212585 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1418791 |
| concepts[0].display_name | Drug discovery |
| concepts[1].id | https://openalex.org/C70721500 |
| concepts[1].level | 1 |
| concepts[1].score | 0.39051762223243713 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[1].display_name | Computational biology |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.36692309379577637 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C86803240 |
| concepts[3].level | 0 |
| concepts[3].score | 0.22072771191596985 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[3].display_name | Biology |
| concepts[4].id | https://openalex.org/C60644358 |
| concepts[4].level | 1 |
| concepts[4].score | 0.21627280116081238 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[4].display_name | Bioinformatics |
| keywords[0].id | https://openalex.org/keywords/drug-discovery |
| keywords[0].score | 0.41158172488212585 |
| keywords[0].display_name | Drug discovery |
| keywords[1].id | https://openalex.org/keywords/computational-biology |
| keywords[1].score | 0.39051762223243713 |
| keywords[1].display_name | Computational biology |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.36692309379577637 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/biology |
| keywords[3].score | 0.22072771191596985 |
| keywords[3].display_name | Biology |
| keywords[4].id | https://openalex.org/keywords/bioinformatics |
| keywords[4].score | 0.21627280116081238 |
| keywords[4].display_name | Bioinformatics |
| language | en |
| locations[0].id | doi:10.1101/2024.10.11.617918 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2024.10.11.617918 |
| locations[1].id | pmid:41306515 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | bioRxiv : the preprint server for biology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/41306515 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5002972643 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-3472-8767 |
| authorships[0].author.display_name | Bengi Ruken Yavuz |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210130649 |
| authorships[0].affiliations[0].raw_affiliation_string | National Cancer Institute at Frederick |
| authorships[0].institutions[0].id | https://openalex.org/I4210130649 |
| authorships[0].institutions[0].ror | https://ror.org/03v6m3209 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I1299022934, https://openalex.org/I1299303238, https://openalex.org/I4210130649, https://openalex.org/I4210140884 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Frederick National Laboratory for Cancer Research |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Bengi Ruken Yavuz |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | National Cancer Institute at Frederick |
| authorships[1].author.id | https://openalex.org/A5071084031 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-9402-4051 |
| authorships[1].author.display_name | Hyunbum Jang |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210130649 |
| authorships[1].affiliations[0].raw_affiliation_string | National Cancer Institute at Frederick |
| authorships[1].institutions[0].id | https://openalex.org/I4210130649 |
| authorships[1].institutions[0].ror | https://ror.org/03v6m3209 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I1299022934, https://openalex.org/I1299303238, https://openalex.org/I4210130649, https://openalex.org/I4210140884 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Frederick National Laboratory for Cancer Research |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Hyunbum Jang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | National Cancer Institute at Frederick |
| authorships[2].author.id | https://openalex.org/A5025357844 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-8115-6415 |
| authorships[2].author.display_name | Ruth Nussinov |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210130649 |
| authorships[2].affiliations[0].raw_affiliation_string | National Cancer Institute at Frederick |
| authorships[2].institutions[0].id | https://openalex.org/I4210130649 |
| authorships[2].institutions[0].ror | https://ror.org/03v6m3209 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I1299022934, https://openalex.org/I1299303238, https://openalex.org/I4210130649, https://openalex.org/I4210140884 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Frederick National Laboratory for Cancer Research |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Ruth Nussinov |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | National Cancer Institute at Frederick |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Anticancer Target Combinations: Network-Informed Signaling-Based Approach to Discovery |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9980999827384949 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W4391375266, https://openalex.org/W2748952813, https://openalex.org/W2390279801, https://openalex.org/W4391913857, https://openalex.org/W2358668433, https://openalex.org/W2013003323, https://openalex.org/W1981076832, https://openalex.org/W2024773645, https://openalex.org/W2001717001, https://openalex.org/W2043147644 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1101/2024.10.11.617918 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2024.10.11.617918 |
| primary_location.id | doi:10.1101/2024.10.11.617918 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2024/10/15/2024.10.11.617918.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2024.10.11.617918 |
| publication_date | 2024-10-15 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W4400438568, https://openalex.org/W2127832858, https://openalex.org/W4379618497, https://openalex.org/W4379056163, https://openalex.org/W1997700247, https://openalex.org/W2795916497, https://openalex.org/W2146448515, https://openalex.org/W2912993305, https://openalex.org/W4318166181, https://openalex.org/W4220776360, https://openalex.org/W4390747020, https://openalex.org/W4385986528, https://openalex.org/W2129242642, https://openalex.org/W3103250275, https://openalex.org/W3089158571, https://openalex.org/W3109453522, https://openalex.org/W3157276232, https://openalex.org/W3170960533, https://openalex.org/W2999142839, https://openalex.org/W3131053290, https://openalex.org/W4313703146, https://openalex.org/W4302287384, https://openalex.org/W4230568286, https://openalex.org/W4317036722, https://openalex.org/W3087175944, https://openalex.org/W3006386963, https://openalex.org/W2939519112, https://openalex.org/W4390911994, https://openalex.org/W4212966936, https://openalex.org/W2608863179, https://openalex.org/W4317553720, https://openalex.org/W4414780401, https://openalex.org/W4398224376, https://openalex.org/W4226055977, https://openalex.org/W4311247426, https://openalex.org/W3164878156, https://openalex.org/W4304892036, https://openalex.org/W4280630832, https://openalex.org/W2124637492, https://openalex.org/W2112868720, https://openalex.org/W2534390616, https://openalex.org/W2578040266, https://openalex.org/W2345356016, https://openalex.org/W4294216483, https://openalex.org/W2036120890, https://openalex.org/W2139132031, https://openalex.org/W2612467560, https://openalex.org/W3020142913, https://openalex.org/W4388721917, https://openalex.org/W3092428270, https://openalex.org/W4396938299, https://openalex.org/W3094801273, https://openalex.org/W3016044799, https://openalex.org/W4305014652, https://openalex.org/W4313382384, https://openalex.org/W2214074259, https://openalex.org/W4389951851, https://openalex.org/W3196809995, https://openalex.org/W4200307603, https://openalex.org/W4388221408, https://openalex.org/W3192034436, https://openalex.org/W4308766741, https://openalex.org/W4391478259, https://openalex.org/W4220699574, https://openalex.org/W2471562228, https://openalex.org/W4378904902, https://openalex.org/W2744377419, https://openalex.org/W2069526613, https://openalex.org/W4399071013, https://openalex.org/W2014457594, https://openalex.org/W2416365238, https://openalex.org/W2144169031, https://openalex.org/W1974024530, https://openalex.org/W2018016654, https://openalex.org/W2923062981, https://openalex.org/W2106947763, https://openalex.org/W2957125530, https://openalex.org/W2096003848, https://openalex.org/W3015686339, https://openalex.org/W4379599707, https://openalex.org/W4304184595, https://openalex.org/W4389451930, https://openalex.org/W4320716025, https://openalex.org/W4392014356, https://openalex.org/W3215936267, https://openalex.org/W3214173072, https://openalex.org/W3042326408, https://openalex.org/W4289711071, https://openalex.org/W2100490225, https://openalex.org/W2598954950, https://openalex.org/W4285498947, https://openalex.org/W3165143074, https://openalex.org/W4305015474, https://openalex.org/W4304695118, https://openalex.org/W4308931322, https://openalex.org/W4389804521, https://openalex.org/W2168133196, https://openalex.org/W4389861133, https://openalex.org/W2050758071, https://openalex.org/W3141372695, https://openalex.org/W2561259078, https://openalex.org/W2119746400, https://openalex.org/W2304920395, https://openalex.org/W1528661249, https://openalex.org/W4291288736, https://openalex.org/W4214853685, https://openalex.org/W2116013203, https://openalex.org/W1969049655, https://openalex.org/W2128339707, https://openalex.org/W4297221746, https://openalex.org/W2015512041, https://openalex.org/W2010601523, https://openalex.org/W4225143246, https://openalex.org/W2086782348, https://openalex.org/W2750588931, https://openalex.org/W4390908283, https://openalex.org/W2130790725, https://openalex.org/W2112947018, https://openalex.org/W2062533676, https://openalex.org/W4324344205, https://openalex.org/W2147566730, https://openalex.org/W1940319397, https://openalex.org/W3004883393, https://openalex.org/W3210558381, https://openalex.org/W2623587811, https://openalex.org/W2410743230, https://openalex.org/W2150318080, https://openalex.org/W4221062827, https://openalex.org/W4323039400, https://openalex.org/W4401854462, https://openalex.org/W3036531306, https://openalex.org/W3114785916, https://openalex.org/W2133198677, https://openalex.org/W3120989527, https://openalex.org/W2149441684, https://openalex.org/W3217485783, https://openalex.org/W2614443510, https://openalex.org/W2782454362, https://openalex.org/W2317662115, https://openalex.org/W2030674117, https://openalex.org/W2145541974, https://openalex.org/W2171707538, https://openalex.org/W1895164037 |
| referenced_works_count | 143 |
| abstract_inverted_index.. | 194 |
| abstract_inverted_index.a | 67, 100 |
| abstract_inverted_index.| | 223, 227, 253 |
| abstract_inverted_index.In | 247 |
| abstract_inverted_index.To | 32, 89 |
| abstract_inverted_index.We | 214 |
| abstract_inverted_index.an | 244 |
| abstract_inverted_index.at | 105 |
| abstract_inverted_index.be | 204 |
| abstract_inverted_index.by | 16, 136, 185, 269 |
| abstract_inverted_index.in | 115, 128, 167, 231 |
| abstract_inverted_index.is | 30, 64, 113, 125 |
| abstract_inverted_index.it | 84, 216 |
| abstract_inverted_index.of | 26, 40, 44 |
| abstract_inverted_index.on | 85, 121 |
| abstract_inverted_index.or | 139, 199 |
| abstract_inverted_index.to | 46, 65, 70, 77, 94, 142, 170, 217 |
| abstract_inverted_index.we | 98 |
| abstract_inverted_index.Our | 62, 111, 146 |
| abstract_inverted_index.The | 24 |
| abstract_inverted_index.aim | 63 |
| abstract_inverted_index.and | 9, 18, 59, 82, 154, 156, 173, 220, 225, 258, 267 |
| abstract_inverted_index.are | 14, 54, 165, 265 |
| abstract_inverted_index.can | 188 |
| abstract_inverted_index.for | 74, 108 |
| abstract_inverted_index.has | 5 |
| abstract_inverted_index.is, | 196 |
| abstract_inverted_index.its | 116, 197 |
| abstract_inverted_index.off | 190 |
| abstract_inverted_index.our | 157, 178, 234, 262 |
| abstract_inverted_index.the | 36, 41, 51, 75, 91, 240 |
| abstract_inverted_index.BRAF | 226, 252 |
| abstract_inverted_index.ESR1 | 222 |
| abstract_inverted_index.That | 195 |
| abstract_inverted_index.This | 124 |
| abstract_inverted_index.been | 22 |
| abstract_inverted_index.cell | 144 |
| abstract_inverted_index.drug | 3, 28, 34, 80, 96, 109, 130, 175, 186 |
| abstract_inverted_index.from | 56, 239 |
| abstract_inverted_index.have | 21 |
| abstract_inverted_index.seen | 6 |
| abstract_inverted_index.shut | 189 |
| abstract_inverted_index.test | 83 |
| abstract_inverted_index.that | 50, 182 |
| abstract_inverted_index.they | 250 |
| abstract_inverted_index.when | 183 |
| abstract_inverted_index.with | 79, 243, 255 |
| abstract_inverted_index.Thus, | 169 |
| abstract_inverted_index.While | 1 |
| abstract_inverted_index.aimed | 104 |
| abstract_inverted_index.based | 120 |
| abstract_inverted_index.being | 119 |
| abstract_inverted_index.could | 209 |
| abstract_inverted_index.data. | 88 |
| abstract_inverted_index.mimic | 171 |
| abstract_inverted_index.modus | 192 |
| abstract_inverted_index.offer | 99 |
| abstract_inverted_index.seeks | 180 |
| abstract_inverted_index.since | 127 |
| abstract_inverted_index.vast. | 31 |
| abstract_inverted_index.would | 203 |
| abstract_inverted_index.PIK3CA | 224, 228, 254 |
| abstract_inverted_index.breast | 219, 232 |
| abstract_inverted_index.cancer | 172 |
| abstract_inverted_index.common | 166 |
| abstract_inverted_index.driver | 163 |
| abstract_inverted_index.lessen | 211 |
| abstract_inverted_index.method | 69 |
| abstract_inverted_index.number | 25 |
| abstract_inverted_index.routes | 141 |
| abstract_inverted_index.select | 33 |
| abstract_inverted_index.single | 12, 95 |
| abstract_inverted_index.temper | 90 |
| abstract_inverted_index.unique | 114 |
| abstract_inverted_index.applied | 215 |
| abstract_inverted_index.blocked | 134 |
| abstract_inverted_index.cancer, | 129, 233, 249 |
| abstract_inverted_index.counter | 174 |
| abstract_inverted_index.develop | 66 |
| abstract_inverted_index.execute | 143 |
| abstract_inverted_index.further | 210 |
| abstract_inverted_index.network | 152 |
| abstract_inverted_index.optimal | 42, 72 |
| abstract_inverted_index.praxis. | 61 |
| abstract_inverted_index.results | 235, 264 |
| abstract_inverted_index.suggest | 236 |
| abstract_inverted_index.through | 207 |
| abstract_inverted_index.ABSTRACT | 273 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Rotating | 206 |
| abstract_inverted_index.advanced | 151 |
| abstract_inverted_index.approach | 149 |
| abstract_inverted_index.blocked. | 205 |
| abstract_inverted_index.bypasses | 133 |
| abstract_inverted_index.clinical | 60 |
| abstract_inverted_index.commonly | 132 |
| abstract_inverted_index.concepts | 153 |
| abstract_inverted_index.discover | 71 |
| abstract_inverted_index.dramatic | 7 |
| abstract_inverted_index.emerging | 212 |
| abstract_inverted_index.expected | 92 |
| abstract_inverted_index.lagging. | 23 |
| abstract_inverted_index.metrics, | 155 |
| abstract_inverted_index.operandi | 193 |
| abstract_inverted_index.parallel | 198 |
| abstract_inverted_index.pathways | 202 |
| abstract_inverted_index.pipeline | 103, 179 |
| abstract_inverted_index.possible | 27 |
| abstract_inverted_index.proteins | 45, 73, 135, 238 |
| abstract_inverted_index.regimen, | 97 |
| abstract_inverted_index.requires | 38 |
| abstract_inverted_index.strategy | 112 |
| abstract_inverted_index.targeted | 184 |
| abstract_inverted_index.wielding | 137 |
| abstract_inverted_index.GRAPHICAL | 272 |
| abstract_inverted_index.co-target | 78, 117, 251 |
| abstract_inverted_index.compiled, | 158 |
| abstract_inverted_index.considers | 53 |
| abstract_inverted_index.discovery | 4 |
| abstract_inverted_index.empirical | 57 |
| abstract_inverted_index.harnesses | 150 |
| abstract_inverted_index.knowledge | 39 |
| abstract_inverted_index.mutations | 164 |
| abstract_inverted_index.pathways. | 123 |
| abstract_inverted_index.primarily | 55 |
| abstract_inverted_index.selecting | 106 |
| abstract_inverted_index.selection | 118 |
| abstract_inverted_index.signaling | 122, 201 |
| abstract_inverted_index.therapies | 13 |
| abstract_inverted_index.validated | 268 |
| abstract_inverted_index.Currently, | 48 |
| abstract_inverted_index.alpelisib, | 256 |
| abstract_inverted_index.anticancer | 2 |
| abstract_inverted_index.available, | 86 |
| abstract_inverted_index.cancer’s | 191 |
| abstract_inverted_index.cetuximab, | 257 |
| abstract_inverted_index.co-target. | 47 |
| abstract_inverted_index.co-targets | 107, 181 |
| abstract_inverted_index.colorectal | 221, 248 |
| abstract_inverted_index.mutations. | 161 |
| abstract_inverted_index.oncologist | 37, 52, 76 |
| abstract_inverted_index.promising, | 266 |
| abstract_inverted_index.resistance | 93, 131, 176 |
| abstract_inverted_index.scenarios, | 177 |
| abstract_inverted_index.sequential | 11 |
| abstract_inverted_index.strategies | 20 |
| abstract_inverted_index.stratified | 102 |
| abstract_inverted_index.subnetwork | 242 |
| abstract_inverted_index.successes, | 10 |
| abstract_inverted_index.Co-existing | 162 |
| abstract_inverted_index.ESR1|PIK3CA | 241 |
| abstract_inverted_index.co-existing | 160 |
| abstract_inverted_index.combination | 43 |
| abstract_inverted_index.encorafenib | 259 |
| abstract_inverted_index.innovations | 8 |
| abstract_inverted_index.resistance, | 17 |
| abstract_inverted_index.resistance. | 168, 213 |
| abstract_inverted_index.significant | 126 |
| abstract_inverted_index.xenografts. | 271 |
| abstract_inverted_index.alternative, | 138 |
| abstract_inverted_index.co-targeting | 237 |
| abstract_inverted_index.combination. | 246, 260 |
| abstract_inverted_index.combinations | 29, 35, 49, 208 |
| abstract_inverted_index.observations | 58 |
| abstract_inverted_index.pipeline’s | 263 |
| abstract_inverted_index.subnetworks. | 229 |
| abstract_inverted_index.time-limited | 15 |
| abstract_inverted_index.Collectively, | 261 |
| abstract_inverted_index.Consistently, | 230 |
| abstract_inverted_index.combinations, | 81, 187 |
| abstract_inverted_index.combinations. | 110 |
| abstract_inverted_index.combinatorial | 19 |
| abstract_inverted_index.complementary | 200 |
| abstract_inverted_index.concept-based | 101 |
| abstract_inverted_index.patient-based | 270 |
| abstract_inverted_index.complementary, | 140 |
| abstract_inverted_index.proliferation. | 145 |
| abstract_inverted_index.patient-derived | 87, 218 |
| abstract_inverted_index.signaling-based | 68, 148 |
| abstract_inverted_index.tissue-specific | 159 |
| abstract_inverted_index.alpelisib-LJM716 | 245 |
| abstract_inverted_index.network-informed | 147 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5002972643 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I4210130649 |
| citation_normalized_percentile.value | 0.25860371 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |