Antifungal mechanism and transcriptome analysis of Bacillomycin D-C16 against Fusarium oxysporum Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.3389/fmicb.2025.1698200
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.3389/fmicb.2025.1698200
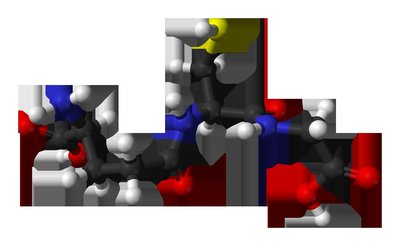
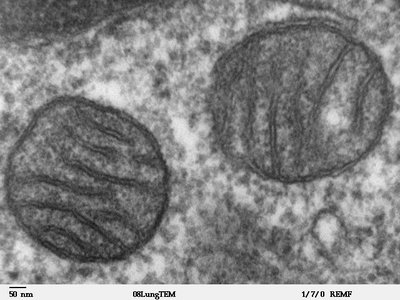
Background Fusarium oxysporum is a globally distributed soil-borne pathogen that causes substantial economic losses in cash crops. Bacillomycin D-C16, a natural antimicrobial lipopeptide produced by Bacillus subtilis , exhibited potent fungicidal activity against F. oxysporum , with a minimum inhibitory concentration (MIC) of 8 mg/L. However, the precise mechanism of its action against F. oxysporum remains uncharacterized. Methods In this study, we employed transmission electron microscopy (TEM) to analyze morphological and ultrastructural alterations in F. oxysporum treated with Bacillomycin D-C16 and RNA-seq profiling combined with biochemical assays to elucidate Bacillomycin D-C16's mode of action against F. oxysporum . Results TEM revealed that Bacillomycin D-C16 induced structural disruption of mitochondria in F. oxysporum . Transcriptome analysis identified 3,370 differentially expressed genes (DEGs) in F. oxysporum , comprising 1,488 up-regulated and 1,882 down-regulated genes. Cluster analysis revealed significant changes in gene expression patterns: DEGs associated with mitochondrial function [including oxidative phosphorylation and citrate cycle (TCA cycle) pathways] were down-regulated, while most DEGs involved in glutathione metabolism were up-regulated. Furthermore, nearly all DEGs related to DNA replication were significantly suppressed. Biochemical assays confirmed these observations: Reduced activities of mitochondrial enzymes [malate dehydrogenase (MDH), isocitrate dehydrogenase (IDH), pyruvate dehydrogenase (PDH), and complexes I–V], decreased mitochondrial membrane potential, and diminished ATP content collectively indicated mitochondrial dysfunction. Depleted glutathione (GSH) levels accompanied by elevated glutathione s-transferase (GST) activity, increased malondialdehyde (MDA) content, and accumulated reactive oxygen species (ROS) confirmed disruptions in glutathione metabolism and oxidative stress. Ultraviolet (UV) absorption spectra, fluorescence spectroscopy, and molecular docking simulations demonstrated Bacillomycin D-C16′s preferential binding to the major groove of DNA, leading to abnormal DNA replication. Conclusions These findings collectively demonstrate that Bacillomycin D-C16 inhibits F. oxysporum growth through multifaceted mechanisms involving transcriptional regulation, mitochondrial impairment, ROS accumulation, and interference with DNA replication.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3389/fmicb.2025.1698200
- https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdf
- OA Status
- gold
- References
- 44
- OpenAlex ID
- https://openalex.org/W7106520121
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W7106520121Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3389/fmicb.2025.1698200Digital Object Identifier
- Title
-
Antifungal mechanism and transcriptome analysis of Bacillomycin D-C16 against Fusarium oxysporumWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-11-25Full publication date if available
- Authors
-
Fu-xing Lin, Yang Jiao, Hua Jiang, Yu Zheng, Shiqin Wu, Li Zhou, Xiangmei Ren, Zhaoxin Lu, Yuanhong LiList of authors in order
- Landing page
-
https://doi.org/10.3389/fmicb.2025.1698200Publisher landing page
- PDF URL
-
https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdfDirect OA link when available
- Concepts
-
Glutathione, Biochemistry, Glutathione disulfide, Transcriptome, Mitochondrion, Succinate dehydrogenase, Biology, Oxidative phosphorylation, Chemistry, Reactive oxygen species, Metabolism, Pyruvate dehydrogenase complex, Citric acid cycle, Mechanism of action, Respiratory chain, Molecular biology, Mitochondrial respiratory chain, Dehydrogenase, Metabolic pathway, Gene expression, Microbiology, Lactate dehydrogenase, Cell biology, Oxidative stress, Superoxide dismutase, Metabolite, Mode of action, Glutathione reductaseTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
44Number of works referenced by this work
Full payload
| id | https://openalex.org/W7106520121 |
|---|---|
| doi | https://doi.org/10.3389/fmicb.2025.1698200 |
| ids.doi | https://doi.org/10.3389/fmicb.2025.1698200 |
| ids.openalex | https://openalex.org/W7106520121 |
| fwci | 0.0 |
| type | article |
| title | Antifungal mechanism and transcriptome analysis of Bacillomycin D-C16 against Fusarium oxysporum |
| biblio.issue | |
| biblio.volume | 16 |
| biblio.last_page | |
| biblio.first_page | |
| is_xpac | False |
| apc_list.value | 2950 |
| apc_list.currency | USD |
| apc_list.value_usd | 2950 |
| apc_paid.value | 2950 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2950 |
| concepts[0].id | https://openalex.org/C538909803 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7641246914863586 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q116907 |
| concepts[0].display_name | Glutathione |
| concepts[1].id | https://openalex.org/C55493867 |
| concepts[1].level | 1 |
| concepts[1].score | 0.6117448210716248 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[1].display_name | Biochemistry |
| concepts[2].id | https://openalex.org/C2779179908 |
| concepts[2].level | 4 |
| concepts[2].score | 0.5136547088623047 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q3324838 |
| concepts[2].display_name | Glutathione disulfide |
| concepts[3].id | https://openalex.org/C162317418 |
| concepts[3].level | 4 |
| concepts[3].score | 0.512954592704773 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q252857 |
| concepts[3].display_name | Transcriptome |
| concepts[4].id | https://openalex.org/C28859421 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5113563537597656 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q39572 |
| concepts[4].display_name | Mitochondrion |
| concepts[5].id | https://openalex.org/C2781330172 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4739111661911011 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q412837 |
| concepts[5].display_name | Succinate dehydrogenase |
| concepts[6].id | https://openalex.org/C86803240 |
| concepts[6].level | 0 |
| concepts[6].score | 0.46750539541244507 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[6].display_name | Biology |
| concepts[7].id | https://openalex.org/C57600042 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4503591060638428 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q227564 |
| concepts[7].display_name | Oxidative phosphorylation |
| concepts[8].id | https://openalex.org/C185592680 |
| concepts[8].level | 0 |
| concepts[8].score | 0.42823731899261475 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[8].display_name | Chemistry |
| concepts[9].id | https://openalex.org/C48349386 |
| concepts[9].level | 2 |
| concepts[9].score | 0.4231501817703247 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q424361 |
| concepts[9].display_name | Reactive oxygen species |
| concepts[10].id | https://openalex.org/C62231903 |
| concepts[10].level | 2 |
| concepts[10].score | 0.3698946535587311 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q1057 |
| concepts[10].display_name | Metabolism |
| concepts[11].id | https://openalex.org/C88099638 |
| concepts[11].level | 3 |
| concepts[11].score | 0.3614576458930969 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q412856 |
| concepts[11].display_name | Pyruvate dehydrogenase complex |
| concepts[12].id | https://openalex.org/C165135838 |
| concepts[12].level | 3 |
| concepts[12].score | 0.341047078371048 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q133895 |
| concepts[12].display_name | Citric acid cycle |
| concepts[13].id | https://openalex.org/C2776120743 |
| concepts[13].level | 3 |
| concepts[13].score | 0.3381282687187195 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q3271540 |
| concepts[13].display_name | Mechanism of action |
| concepts[14].id | https://openalex.org/C3020377403 |
| concepts[14].level | 3 |
| concepts[14].score | 0.3347301185131073 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q211016 |
| concepts[14].display_name | Respiratory chain |
| concepts[15].id | https://openalex.org/C153911025 |
| concepts[15].level | 1 |
| concepts[15].score | 0.32540908455848694 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7202 |
| concepts[15].display_name | Molecular biology |
| concepts[16].id | https://openalex.org/C3020761558 |
| concepts[16].level | 3 |
| concepts[16].score | 0.3086893558502197 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q211016 |
| concepts[16].display_name | Mitochondrial respiratory chain |
| concepts[17].id | https://openalex.org/C2776317432 |
| concepts[17].level | 3 |
| concepts[17].score | 0.3014279901981354 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q412547 |
| concepts[17].display_name | Dehydrogenase |
| concepts[18].id | https://openalex.org/C192989942 |
| concepts[18].level | 3 |
| concepts[18].score | 0.29991450905799866 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q68685 |
| concepts[18].display_name | Metabolic pathway |
| concepts[19].id | https://openalex.org/C150194340 |
| concepts[19].level | 3 |
| concepts[19].score | 0.29215776920318604 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[19].display_name | Gene expression |
| concepts[20].id | https://openalex.org/C89423630 |
| concepts[20].level | 1 |
| concepts[20].score | 0.2902611196041107 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[20].display_name | Microbiology |
| concepts[21].id | https://openalex.org/C2777318727 |
| concepts[21].level | 3 |
| concepts[21].score | 0.27842190861701965 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q409464 |
| concepts[21].display_name | Lactate dehydrogenase |
| concepts[22].id | https://openalex.org/C95444343 |
| concepts[22].level | 1 |
| concepts[22].score | 0.2758534252643585 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[22].display_name | Cell biology |
| concepts[23].id | https://openalex.org/C2776151105 |
| concepts[23].level | 2 |
| concepts[23].score | 0.27442821860313416 |
| concepts[23].wikidata | https://www.wikidata.org/wiki/Q898814 |
| concepts[23].display_name | Oxidative stress |
| concepts[24].id | https://openalex.org/C2775838275 |
| concepts[24].level | 3 |
| concepts[24].score | 0.2670772671699524 |
| concepts[24].wikidata | https://www.wikidata.org/wiki/Q410776 |
| concepts[24].display_name | Superoxide dismutase |
| concepts[25].id | https://openalex.org/C2777477808 |
| concepts[25].level | 2 |
| concepts[25].score | 0.2631851136684418 |
| concepts[25].wikidata | https://www.wikidata.org/wiki/Q407595 |
| concepts[25].display_name | Metabolite |
| concepts[26].id | https://openalex.org/C121587506 |
| concepts[26].level | 2 |
| concepts[26].score | 0.2606469392776489 |
| concepts[26].wikidata | https://www.wikidata.org/wiki/Q6888113 |
| concepts[26].display_name | Mode of action |
| concepts[27].id | https://openalex.org/C165069038 |
| concepts[27].level | 5 |
| concepts[27].score | 0.2605791389942169 |
| concepts[27].wikidata | https://www.wikidata.org/wiki/Q420930 |
| concepts[27].display_name | Glutathione reductase |
| keywords[0].id | https://openalex.org/keywords/glutathione |
| keywords[0].score | 0.7641246914863586 |
| keywords[0].display_name | Glutathione |
| keywords[1].id | https://openalex.org/keywords/glutathione-disulfide |
| keywords[1].score | 0.5136547088623047 |
| keywords[1].display_name | Glutathione disulfide |
| keywords[2].id | https://openalex.org/keywords/transcriptome |
| keywords[2].score | 0.512954592704773 |
| keywords[2].display_name | Transcriptome |
| keywords[3].id | https://openalex.org/keywords/mitochondrion |
| keywords[3].score | 0.5113563537597656 |
| keywords[3].display_name | Mitochondrion |
| keywords[4].id | https://openalex.org/keywords/succinate-dehydrogenase |
| keywords[4].score | 0.4739111661911011 |
| keywords[4].display_name | Succinate dehydrogenase |
| keywords[5].id | https://openalex.org/keywords/oxidative-phosphorylation |
| keywords[5].score | 0.4503591060638428 |
| keywords[5].display_name | Oxidative phosphorylation |
| keywords[6].id | https://openalex.org/keywords/reactive-oxygen-species |
| keywords[6].score | 0.4231501817703247 |
| keywords[6].display_name | Reactive oxygen species |
| keywords[7].id | https://openalex.org/keywords/metabolism |
| keywords[7].score | 0.3698946535587311 |
| keywords[7].display_name | Metabolism |
| keywords[8].id | https://openalex.org/keywords/pyruvate-dehydrogenase-complex |
| keywords[8].score | 0.3614576458930969 |
| keywords[8].display_name | Pyruvate dehydrogenase complex |
| language | en |
| locations[0].id | doi:10.3389/fmicb.2025.1698200 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2484704948 |
| locations[0].source.issn | 1664-302X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1664-302X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Frontiers in Microbiology |
| locations[0].source.host_organization | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_name | Frontiers Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320527 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Frontiers in Microbiology |
| locations[0].landing_page_url | https://doi.org/10.3389/fmicb.2025.1698200 |
| locations[1].id | pmh:oai:doaj.org/article:81dae00b2bd04025acc890637b7f52d5 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306401280 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].source.host_organization_lineage | |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | article |
| locations[1].license_id | |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | Frontiers in Microbiology, Vol 16 (2025) |
| locations[1].landing_page_url | https://doaj.org/article/81dae00b2bd04025acc890637b7f52d5 |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A2344518169 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-8699-1313 |
| authorships[0].author.display_name | Fu-xing Lin |
| authorships[0].countries | CN |
| authorships[0].institutions[0].id | https://openalex.org/I177388780 |
| authorships[0].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Xuzhou Medical College |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Fuxing Lin |
| authorships[0].is_corresponding | True |
| authorships[1].author.id | https://openalex.org/A2037978905 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-5603-0813 |
| authorships[1].author.display_name | Yang Jiao |
| authorships[1].countries | CN |
| authorships[1].institutions[0].id | https://openalex.org/I177388780 |
| authorships[1].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Xuzhou Medical College |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Yang Jiao |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A2103193790 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-5556-5517 |
| authorships[2].author.display_name | Hua Jiang |
| authorships[2].countries | CN |
| authorships[2].institutions[0].id | https://openalex.org/I177388780 |
| authorships[2].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Xuzhou Medical College |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Hua Jiang |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A2079030413 |
| authorships[3].author.orcid | https://orcid.org/0000-0001-5036-4310 |
| authorships[3].author.display_name | Yu Zheng |
| authorships[3].countries | CN |
| authorships[3].institutions[0].id | https://openalex.org/I177388780 |
| authorships[3].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Xuzhou Medical College |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Yu Zheng |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A2292279470 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Shiqin Wu |
| authorships[4].countries | CN |
| authorships[4].institutions[0].id | https://openalex.org/I177388780 |
| authorships[4].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Xuzhou Medical College |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Shiqin Wu |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A2019114847 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-5214-8232 |
| authorships[5].author.display_name | Li Zhou |
| authorships[5].countries | CN |
| authorships[5].institutions[0].id | https://openalex.org/I177388780 |
| authorships[5].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | Xuzhou Medical College |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Li Zhou |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A2416649155 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-4446-3635 |
| authorships[6].author.display_name | Xiangmei Ren |
| authorships[6].countries | CN |
| authorships[6].institutions[0].id | https://openalex.org/I177388780 |
| authorships[6].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[6].institutions[0].country_code | CN |
| authorships[6].institutions[0].display_name | Xuzhou Medical College |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Xiangmei Ren |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A2101761990 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-9288-7419 |
| authorships[7].author.display_name | Zhaoxin Lu |
| authorships[7].countries | CN |
| authorships[7].institutions[0].id | https://openalex.org/I119454577 |
| authorships[7].institutions[0].ror | https://ror.org/https://ror.org/05td3s095 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I119454577 |
| authorships[7].institutions[0].country_code | CN |
| authorships[7].institutions[0].display_name | Nanjing Agricultural University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Zhaoxin Lu |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A2105451704 |
| authorships[8].author.orcid | https://orcid.org/0000-0001-5526-4706 |
| authorships[8].author.display_name | Yuanhong Li |
| authorships[8].countries | CN |
| authorships[8].institutions[0].id | https://openalex.org/I177388780 |
| authorships[8].institutions[0].ror | https://ror.org/https://ror.org/04fe7hy80 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I177388780 |
| authorships[8].institutions[0].country_code | CN |
| authorships[8].institutions[0].display_name | Xuzhou Medical College |
| authorships[8].author_position | last |
| authorships[8].raw_author_name | Yuanhong Li |
| authorships[8].is_corresponding | False |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-11-25T00:00:00 |
| display_name | Antifungal mechanism and transcriptome analysis of Bacillomycin D-C16 against Fusarium oxysporum |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-27T01:12:40.094763 |
| primary_topic | |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.3389/fmicb.2025.1698200 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2484704948 |
| best_oa_location.source.issn | 1664-302X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1664-302X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Frontiers in Microbiology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_name | Frontiers Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Frontiers in Microbiology |
| best_oa_location.landing_page_url | https://doi.org/10.3389/fmicb.2025.1698200 |
| primary_location.id | doi:10.3389/fmicb.2025.1698200 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2484704948 |
| primary_location.source.issn | 1664-302X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1664-302X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Frontiers in Microbiology |
| primary_location.source.host_organization | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_name | Frontiers Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://public-pages-files-2025.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1698200/pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Frontiers in Microbiology |
| primary_location.landing_page_url | https://doi.org/10.3389/fmicb.2025.1698200 |
| publication_date | 2025-11-25 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W2106130068, https://openalex.org/W4384298949, https://openalex.org/W4224115038, https://openalex.org/W4393192296, https://openalex.org/W3134912752, https://openalex.org/W4297373163, https://openalex.org/W4395961165, https://openalex.org/W2737732597, https://openalex.org/W2899093583, https://openalex.org/W3196959712, https://openalex.org/W4400905701, https://openalex.org/W2983062612, https://openalex.org/W2098530863, https://openalex.org/W4388996308, https://openalex.org/W4316499269, https://openalex.org/W2889388610, https://openalex.org/W4200162499, https://openalex.org/W2924119290, https://openalex.org/W2981152601, https://openalex.org/W4411325449, https://openalex.org/W2107277218, https://openalex.org/W2997046468, https://openalex.org/W3109355298, https://openalex.org/W2794060187, https://openalex.org/W3174895520, https://openalex.org/W1228044642, https://openalex.org/W4408157855, https://openalex.org/W4312052458, https://openalex.org/W3201911427, https://openalex.org/W4297349551, https://openalex.org/W4399173206, https://openalex.org/W2148411571, https://openalex.org/W4392158382, https://openalex.org/W3007446909, https://openalex.org/W2027847170, https://openalex.org/W4391683178, https://openalex.org/W4409542832, https://openalex.org/W4380081806, https://openalex.org/W4394817699, https://openalex.org/W4388406093, https://openalex.org/W2768314152, https://openalex.org/W4361268172, https://openalex.org/W2626993113, https://openalex.org/W4281742097 |
| referenced_works_count | 44 |
| abstract_inverted_index., | 27, 35, 124 |
| abstract_inverted_index.. | 97, 112 |
| abstract_inverted_index.8 | 43 |
| abstract_inverted_index.a | 4, 19, 37 |
| abstract_inverted_index.F. | 33, 53, 74, 95, 110, 122, 275 |
| abstract_inverted_index.In | 58 |
| abstract_inverted_index.by | 24, 216 |
| abstract_inverted_index.in | 14, 73, 109, 121, 137, 161, 234 |
| abstract_inverted_index.is | 3 |
| abstract_inverted_index.of | 42, 49, 92, 107, 184, 259 |
| abstract_inverted_index.to | 67, 87, 171, 255, 262 |
| abstract_inverted_index.we | 61 |
| abstract_inverted_index.ATP | 205 |
| abstract_inverted_index.DNA | 172, 264, 291 |
| abstract_inverted_index.ROS | 286 |
| abstract_inverted_index.TEM | 99 |
| abstract_inverted_index.all | 168 |
| abstract_inverted_index.and | 70, 80, 128, 149, 196, 203, 226, 237, 246, 288 |
| abstract_inverted_index.its | 50 |
| abstract_inverted_index.the | 46, 256 |
| abstract_inverted_index.(TCA | 152 |
| abstract_inverted_index.(UV) | 241 |
| abstract_inverted_index.DEGs | 141, 159, 169 |
| abstract_inverted_index.DNA, | 260 |
| abstract_inverted_index.cash | 15 |
| abstract_inverted_index.gene | 138 |
| abstract_inverted_index.mode | 91 |
| abstract_inverted_index.most | 158 |
| abstract_inverted_index.that | 9, 101, 271 |
| abstract_inverted_index.this | 59 |
| abstract_inverted_index.were | 155, 164, 174 |
| abstract_inverted_index.with | 36, 77, 84, 143, 290 |
| abstract_inverted_index.(GSH) | 213 |
| abstract_inverted_index.(GST) | 220 |
| abstract_inverted_index.(MDA) | 224 |
| abstract_inverted_index.(MIC) | 41 |
| abstract_inverted_index.(ROS) | 231 |
| abstract_inverted_index.(TEM) | 66 |
| abstract_inverted_index.1,488 | 126 |
| abstract_inverted_index.1,882 | 129 |
| abstract_inverted_index.3,370 | 116 |
| abstract_inverted_index.D-C16 | 79, 103, 273 |
| abstract_inverted_index.These | 267 |
| abstract_inverted_index.cycle | 151 |
| abstract_inverted_index.genes | 119 |
| abstract_inverted_index.major | 257 |
| abstract_inverted_index.mg/L. | 44 |
| abstract_inverted_index.these | 180 |
| abstract_inverted_index.while | 157 |
| abstract_inverted_index.(DEGs) | 120 |
| abstract_inverted_index.(IDH), | 192 |
| abstract_inverted_index.(MDH), | 189 |
| abstract_inverted_index.(PDH), | 195 |
| abstract_inverted_index.D-C16, | 18 |
| abstract_inverted_index.action | 51, 93 |
| abstract_inverted_index.assays | 86, 178 |
| abstract_inverted_index.causes | 10 |
| abstract_inverted_index.crops. | 16 |
| abstract_inverted_index.cycle) | 153 |
| abstract_inverted_index.genes. | 131 |
| abstract_inverted_index.groove | 258 |
| abstract_inverted_index.growth | 277 |
| abstract_inverted_index.levels | 214 |
| abstract_inverted_index.losses | 13 |
| abstract_inverted_index.nearly | 167 |
| abstract_inverted_index.oxygen | 229 |
| abstract_inverted_index.potent | 29 |
| abstract_inverted_index.study, | 60 |
| abstract_inverted_index.Cluster | 132 |
| abstract_inverted_index.D-C16's | 90 |
| abstract_inverted_index.I–V], | 198 |
| abstract_inverted_index.Methods | 57 |
| abstract_inverted_index.RNA-seq | 81 |
| abstract_inverted_index.Reduced | 182 |
| abstract_inverted_index.Results | 98 |
| abstract_inverted_index.[malate | 187 |
| abstract_inverted_index.against | 32, 52, 94 |
| abstract_inverted_index.analyze | 68 |
| abstract_inverted_index.binding | 254 |
| abstract_inverted_index.changes | 136 |
| abstract_inverted_index.citrate | 150 |
| abstract_inverted_index.content | 206 |
| abstract_inverted_index.docking | 248 |
| abstract_inverted_index.enzymes | 186 |
| abstract_inverted_index.induced | 104 |
| abstract_inverted_index.leading | 261 |
| abstract_inverted_index.minimum | 38 |
| abstract_inverted_index.natural | 20 |
| abstract_inverted_index.precise | 47 |
| abstract_inverted_index.related | 170 |
| abstract_inverted_index.remains | 55 |
| abstract_inverted_index.species | 230 |
| abstract_inverted_index.stress. | 239 |
| abstract_inverted_index.through | 278 |
| abstract_inverted_index.treated | 76 |
| abstract_inverted_index.Bacillus | 25 |
| abstract_inverted_index.Depleted | 211 |
| abstract_inverted_index.Fusarium | 1 |
| abstract_inverted_index.However, | 45 |
| abstract_inverted_index.abnormal | 263 |
| abstract_inverted_index.activity | 31 |
| abstract_inverted_index.analysis | 114, 133 |
| abstract_inverted_index.combined | 83 |
| abstract_inverted_index.content, | 225 |
| abstract_inverted_index.economic | 12 |
| abstract_inverted_index.electron | 64 |
| abstract_inverted_index.elevated | 217 |
| abstract_inverted_index.employed | 62 |
| abstract_inverted_index.findings | 268 |
| abstract_inverted_index.function | 145 |
| abstract_inverted_index.globally | 5 |
| abstract_inverted_index.inhibits | 274 |
| abstract_inverted_index.involved | 160 |
| abstract_inverted_index.membrane | 201 |
| abstract_inverted_index.pathogen | 8 |
| abstract_inverted_index.produced | 23 |
| abstract_inverted_index.pyruvate | 193 |
| abstract_inverted_index.reactive | 228 |
| abstract_inverted_index.revealed | 100, 134 |
| abstract_inverted_index.spectra, | 243 |
| abstract_inverted_index.subtilis | 26 |
| abstract_inverted_index.D-C16′s | 252 |
| abstract_inverted_index.activity, | 221 |
| abstract_inverted_index.complexes | 197 |
| abstract_inverted_index.confirmed | 179, 232 |
| abstract_inverted_index.decreased | 199 |
| abstract_inverted_index.elucidate | 88 |
| abstract_inverted_index.exhibited | 28 |
| abstract_inverted_index.expressed | 118 |
| abstract_inverted_index.increased | 222 |
| abstract_inverted_index.indicated | 208 |
| abstract_inverted_index.involving | 281 |
| abstract_inverted_index.mechanism | 48 |
| abstract_inverted_index.molecular | 247 |
| abstract_inverted_index.oxidative | 147, 238 |
| abstract_inverted_index.oxysporum | 2, 34, 54, 75, 96, 111, 123, 276 |
| abstract_inverted_index.pathways] | 154 |
| abstract_inverted_index.patterns: | 140 |
| abstract_inverted_index.profiling | 82 |
| abstract_inverted_index.Background | 0 |
| abstract_inverted_index.[including | 146 |
| abstract_inverted_index.absorption | 242 |
| abstract_inverted_index.activities | 183 |
| abstract_inverted_index.associated | 142 |
| abstract_inverted_index.comprising | 125 |
| abstract_inverted_index.diminished | 204 |
| abstract_inverted_index.disruption | 106 |
| abstract_inverted_index.expression | 139 |
| abstract_inverted_index.fungicidal | 30 |
| abstract_inverted_index.identified | 115 |
| abstract_inverted_index.inhibitory | 39 |
| abstract_inverted_index.isocitrate | 190 |
| abstract_inverted_index.mechanisms | 280 |
| abstract_inverted_index.metabolism | 163, 236 |
| abstract_inverted_index.microscopy | 65 |
| abstract_inverted_index.potential, | 202 |
| abstract_inverted_index.soil-borne | 7 |
| abstract_inverted_index.structural | 105 |
| abstract_inverted_index.Biochemical | 177 |
| abstract_inverted_index.Conclusions | 266 |
| abstract_inverted_index.Ultraviolet | 240 |
| abstract_inverted_index.accompanied | 215 |
| abstract_inverted_index.accumulated | 227 |
| abstract_inverted_index.alterations | 72 |
| abstract_inverted_index.biochemical | 85 |
| abstract_inverted_index.demonstrate | 270 |
| abstract_inverted_index.disruptions | 233 |
| abstract_inverted_index.distributed | 6 |
| abstract_inverted_index.glutathione | 162, 212, 218, 235 |
| abstract_inverted_index.impairment, | 285 |
| abstract_inverted_index.lipopeptide | 22 |
| abstract_inverted_index.regulation, | 283 |
| abstract_inverted_index.replication | 173 |
| abstract_inverted_index.significant | 135 |
| abstract_inverted_index.simulations | 249 |
| abstract_inverted_index.substantial | 11 |
| abstract_inverted_index.suppressed. | 176 |
| abstract_inverted_index.Bacillomycin | 17, 78, 89, 102, 251, 272 |
| abstract_inverted_index.Furthermore, | 166 |
| abstract_inverted_index.collectively | 207, 269 |
| abstract_inverted_index.demonstrated | 250 |
| abstract_inverted_index.dysfunction. | 210 |
| abstract_inverted_index.fluorescence | 244 |
| abstract_inverted_index.interference | 289 |
| abstract_inverted_index.mitochondria | 108 |
| abstract_inverted_index.multifaceted | 279 |
| abstract_inverted_index.preferential | 253 |
| abstract_inverted_index.replication. | 265, 292 |
| abstract_inverted_index.transmission | 63 |
| abstract_inverted_index.up-regulated | 127 |
| abstract_inverted_index.Transcriptome | 113 |
| abstract_inverted_index.accumulation, | 287 |
| abstract_inverted_index.antimicrobial | 21 |
| abstract_inverted_index.concentration | 40 |
| abstract_inverted_index.dehydrogenase | 188, 191, 194 |
| abstract_inverted_index.mitochondrial | 144, 185, 200, 209, 284 |
| abstract_inverted_index.morphological | 69 |
| abstract_inverted_index.observations: | 181 |
| abstract_inverted_index.s-transferase | 219 |
| abstract_inverted_index.significantly | 175 |
| abstract_inverted_index.spectroscopy, | 245 |
| abstract_inverted_index.up-regulated. | 165 |
| abstract_inverted_index.differentially | 117 |
| abstract_inverted_index.down-regulated | 130 |
| abstract_inverted_index.down-regulated, | 156 |
| abstract_inverted_index.malondialdehyde | 223 |
| abstract_inverted_index.phosphorylation | 148 |
| abstract_inverted_index.transcriptional | 282 |
| abstract_inverted_index.ultrastructural | 71 |
| abstract_inverted_index.uncharacterized. | 56 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 9 |
| citation_normalized_percentile |