Association of body index with fecal microbiome in children cohorts with ethnic–geographic factor interaction: accurately using a Bayesian zero-inflated negative binomial regression model Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1128/msystems.01345-24
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1128/msystems.01345-24
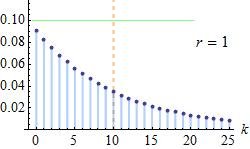
The exponential growth of high-throughput sequencing (HTS) data on the microbial communities presents researchers with an unparalleled opportunity to delve deeper into the association of microorganisms with host phenotype. However, this growth also poses a challenge, as microbial data are complex, sparse, discrete, and prone to zero inflation. Herein, by utilizing 10 distinct counting models for analyzing simulated data, we proposed an innovative Bayesian zero-inflated negative binomial (ZINB) regression model that is capable of identifying differentially abundant taxa associated with distinctive host phenotypes and quantifying the effects of covariates on these taxa. Our proposed model exhibits excellent accuracy compared with conventional Hurdle and INLA models, especially in scenarios characterized by inflation and overdispersion. Moreover, we confirm that dispersion parameters significantly affect the accuracy of model results, with defects gradually alleviating as the number of analyzed samples increases. Subsequently applying our model to amplicon data in real multi-ethnic children cohort, we found that only a subset of taxa were identified as having zero inflation in real data, suggesting that the prevailing understanding and processing of microbial count data in most previous microbiome studies were overly dogmatic. In practice, our pipeline of integrating bacterial differential abundance in microbiome data and relevant covariates is effective and feasible. Taken together, our method is expected to be extended to the microbiota studies of various multi-cohort populations. IMPORTANCE The microbiome is closely associated with physical indicators of the body, such as height, weight, age and BMI, which can be used as measures of human health. Accurately identifying which taxa in the microbiome are closely related to indicators of physical development is valuable as microbial markers of regional child growth trajectory. Zero-inflated negative binomial (ZINB) model, a type of Bayesian generalized linear model, can be effectively modeled in complex biological systems. We present an innovative ZINB regression model that is capable of identifying differentially abundant taxa associated with distinctive host phenotypes and quantifying the effects of covariates on these taxa, and demonstrate that its accuracy is superior to traditional Hurdle and INLA models. Our pipeline of integrating bacterial differential abundance in microbiome data and relevant covariates is effective and feasible.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1128/msystems.01345-24
- OA Status
- gold
- Cited By
- 1
- References
- 70
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4404589389
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4404589389Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1128/msystems.01345-24Digital Object Identifier
- Title
-
Association of body index with fecal microbiome in children cohorts with ethnic–geographic factor interaction: accurately using a Bayesian zero-inflated negative binomial regression modelWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-11-21Full publication date if available
- Authors
-
Jian Huang, Yanzhuan Lu, Fengwei Tian, Yongqing NiList of authors in order
- Landing page
-
https://doi.org/10.1128/msystems.01345-24Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1128/msystems.01345-24Direct OA link when available
- Concepts
-
Overdispersion, Negative binomial distribution, Count data, Microbiome, Covariate, Statistics, Bayes factor, Biology, Regression, Poisson distribution, Bayesian probability, Econometrics, Bayesian inference, Mathematics, BioinformaticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
70Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4404589389 |
|---|---|
| doi | https://doi.org/10.1128/msystems.01345-24 |
| ids.doi | https://doi.org/10.1128/msystems.01345-24 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39570024 |
| ids.openalex | https://openalex.org/W4404589389 |
| fwci | 0.48024723 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D006801 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Humans |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D001499 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Bayes Theorem |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D002648 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Child |
| mesh[3].qualifier_ui | Q000382 |
| mesh[3].descriptor_ui | D005243 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | microbiology |
| mesh[3].descriptor_name | Feces |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D015992 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Body Mass Index |
| mesh[5].qualifier_ui | Q000235 |
| mesh[5].descriptor_ui | D000069196 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | genetics |
| mesh[5].descriptor_name | Gastrointestinal Microbiome |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D005260 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Female |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D008297 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Male |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D002675 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Child, Preschool |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D015331 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Cohort Studies |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D005006 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Ethnicity |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D001419 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | Bacteria |
| mesh[12].qualifier_ui | Q000145 |
| mesh[12].descriptor_ui | D001419 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | classification |
| mesh[12].descriptor_name | Bacteria |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D012044 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Regression Analysis |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D006801 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Humans |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D001499 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Bayes Theorem |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D002648 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Child |
| mesh[17].qualifier_ui | Q000382 |
| mesh[17].descriptor_ui | D005243 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | microbiology |
| mesh[17].descriptor_name | Feces |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D015992 |
| mesh[18].is_major_topic | True |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Body Mass Index |
| mesh[19].qualifier_ui | Q000235 |
| mesh[19].descriptor_ui | D000069196 |
| mesh[19].is_major_topic | True |
| mesh[19].qualifier_name | genetics |
| mesh[19].descriptor_name | Gastrointestinal Microbiome |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D005260 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Female |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D008297 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Male |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D002675 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Child, Preschool |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D015331 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Cohort Studies |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D005006 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Ethnicity |
| mesh[25].qualifier_ui | Q000235 |
| mesh[25].descriptor_ui | D001419 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | genetics |
| mesh[25].descriptor_name | Bacteria |
| mesh[26].qualifier_ui | Q000145 |
| mesh[26].descriptor_ui | D001419 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | classification |
| mesh[26].descriptor_name | Bacteria |
| mesh[27].qualifier_ui | |
| mesh[27].descriptor_ui | D012044 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | |
| mesh[27].descriptor_name | Regression Analysis |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D006801 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Humans |
| mesh[29].qualifier_ui | |
| mesh[29].descriptor_ui | D001499 |
| mesh[29].is_major_topic | True |
| mesh[29].qualifier_name | |
| mesh[29].descriptor_name | Bayes Theorem |
| mesh[30].qualifier_ui | |
| mesh[30].descriptor_ui | D002648 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | |
| mesh[30].descriptor_name | Child |
| mesh[31].qualifier_ui | Q000382 |
| mesh[31].descriptor_ui | D005243 |
| mesh[31].is_major_topic | True |
| mesh[31].qualifier_name | microbiology |
| mesh[31].descriptor_name | Feces |
| mesh[32].qualifier_ui | |
| mesh[32].descriptor_ui | D015992 |
| mesh[32].is_major_topic | True |
| mesh[32].qualifier_name | |
| mesh[32].descriptor_name | Body Mass Index |
| mesh[33].qualifier_ui | Q000235 |
| mesh[33].descriptor_ui | D000069196 |
| mesh[33].is_major_topic | True |
| mesh[33].qualifier_name | genetics |
| mesh[33].descriptor_name | Gastrointestinal Microbiome |
| mesh[34].qualifier_ui | |
| mesh[34].descriptor_ui | D005260 |
| mesh[34].is_major_topic | False |
| mesh[34].qualifier_name | |
| mesh[34].descriptor_name | Female |
| mesh[35].qualifier_ui | |
| mesh[35].descriptor_ui | D008297 |
| mesh[35].is_major_topic | False |
| mesh[35].qualifier_name | |
| mesh[35].descriptor_name | Male |
| mesh[36].qualifier_ui | |
| mesh[36].descriptor_ui | D002675 |
| mesh[36].is_major_topic | False |
| mesh[36].qualifier_name | |
| mesh[36].descriptor_name | Child, Preschool |
| mesh[37].qualifier_ui | |
| mesh[37].descriptor_ui | D015331 |
| mesh[37].is_major_topic | False |
| mesh[37].qualifier_name | |
| mesh[37].descriptor_name | Cohort Studies |
| mesh[38].qualifier_ui | |
| mesh[38].descriptor_ui | D005006 |
| mesh[38].is_major_topic | False |
| mesh[38].qualifier_name | |
| mesh[38].descriptor_name | Ethnicity |
| mesh[39].qualifier_ui | Q000235 |
| mesh[39].descriptor_ui | D001419 |
| mesh[39].is_major_topic | False |
| mesh[39].qualifier_name | genetics |
| mesh[39].descriptor_name | Bacteria |
| mesh[40].qualifier_ui | Q000145 |
| mesh[40].descriptor_ui | D001419 |
| mesh[40].is_major_topic | False |
| mesh[40].qualifier_name | classification |
| mesh[40].descriptor_name | Bacteria |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D012044 |
| mesh[41].is_major_topic | False |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | Regression Analysis |
| mesh[42].qualifier_ui | |
| mesh[42].descriptor_ui | D006801 |
| mesh[42].is_major_topic | False |
| mesh[42].qualifier_name | |
| mesh[42].descriptor_name | Humans |
| mesh[43].qualifier_ui | |
| mesh[43].descriptor_ui | D001499 |
| mesh[43].is_major_topic | True |
| mesh[43].qualifier_name | |
| mesh[43].descriptor_name | Bayes Theorem |
| mesh[44].qualifier_ui | |
| mesh[44].descriptor_ui | D002648 |
| mesh[44].is_major_topic | False |
| mesh[44].qualifier_name | |
| mesh[44].descriptor_name | Child |
| mesh[45].qualifier_ui | Q000382 |
| mesh[45].descriptor_ui | D005243 |
| mesh[45].is_major_topic | True |
| mesh[45].qualifier_name | microbiology |
| mesh[45].descriptor_name | Feces |
| mesh[46].qualifier_ui | |
| mesh[46].descriptor_ui | D015992 |
| mesh[46].is_major_topic | True |
| mesh[46].qualifier_name | |
| mesh[46].descriptor_name | Body Mass Index |
| mesh[47].qualifier_ui | Q000235 |
| mesh[47].descriptor_ui | D000069196 |
| mesh[47].is_major_topic | True |
| mesh[47].qualifier_name | genetics |
| mesh[47].descriptor_name | Gastrointestinal Microbiome |
| mesh[48].qualifier_ui | |
| mesh[48].descriptor_ui | D005260 |
| mesh[48].is_major_topic | False |
| mesh[48].qualifier_name | |
| mesh[48].descriptor_name | Female |
| mesh[49].qualifier_ui | |
| mesh[49].descriptor_ui | D008297 |
| mesh[49].is_major_topic | False |
| mesh[49].qualifier_name | |
| mesh[49].descriptor_name | Male |
| type | article |
| title | Association of body index with fecal microbiome in children cohorts with ethnic–geographic factor interaction: accurately using a Bayesian zero-inflated negative binomial regression model |
| biblio.issue | 12 |
| biblio.volume | 9 |
| biblio.last_page | e0134524 |
| biblio.first_page | e0134524 |
| topics[0].id | https://openalex.org/T10066 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Gut microbiota and health |
| topics[1].id | https://openalex.org/T11610 |
| topics[1].field.id | https://openalex.org/fields/36 |
| topics[1].field.display_name | Health Professions |
| topics[1].score | 0.9596999883651733 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/3600 |
| topics[1].subfield.display_name | General Health Professions |
| topics[1].display_name | Food Security and Health in Diverse Populations |
| topics[2].id | https://openalex.org/T10351 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.958899974822998 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2713 |
| topics[2].subfield.display_name | Epidemiology |
| topics[2].display_name | Liver Disease Diagnosis and Treatment |
| is_xpac | False |
| apc_list.value | 3500 |
| apc_list.currency | USD |
| apc_list.value_usd | 3500 |
| apc_paid.value | 3500 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 3500 |
| concepts[0].id | https://openalex.org/C117236510 |
| concepts[0].level | 4 |
| concepts[0].score | 0.7616919279098511 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q7113620 |
| concepts[0].display_name | Overdispersion |
| concepts[1].id | https://openalex.org/C199335787 |
| concepts[1].level | 3 |
| concepts[1].score | 0.7409213781356812 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q743364 |
| concepts[1].display_name | Negative binomial distribution |
| concepts[2].id | https://openalex.org/C33643355 |
| concepts[2].level | 3 |
| concepts[2].score | 0.584478497505188 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q5176731 |
| concepts[2].display_name | Count data |
| concepts[3].id | https://openalex.org/C143121216 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5826258063316345 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1330402 |
| concepts[3].display_name | Microbiome |
| concepts[4].id | https://openalex.org/C119043178 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5821583867073059 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q320723 |
| concepts[4].display_name | Covariate |
| concepts[5].id | https://openalex.org/C105795698 |
| concepts[5].level | 1 |
| concepts[5].score | 0.5059203505516052 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[5].display_name | Statistics |
| concepts[6].id | https://openalex.org/C142291917 |
| concepts[6].level | 4 |
| concepts[6].score | 0.49013885855674744 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q4165283 |
| concepts[6].display_name | Bayes factor |
| concepts[7].id | https://openalex.org/C86803240 |
| concepts[7].level | 0 |
| concepts[7].score | 0.46630609035491943 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[7].display_name | Biology |
| concepts[8].id | https://openalex.org/C83546350 |
| concepts[8].level | 2 |
| concepts[8].score | 0.46408554911613464 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q1139051 |
| concepts[8].display_name | Regression |
| concepts[9].id | https://openalex.org/C100906024 |
| concepts[9].level | 2 |
| concepts[9].score | 0.4227152466773987 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q205692 |
| concepts[9].display_name | Poisson distribution |
| concepts[10].id | https://openalex.org/C107673813 |
| concepts[10].level | 2 |
| concepts[10].score | 0.4134995937347412 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q812534 |
| concepts[10].display_name | Bayesian probability |
| concepts[11].id | https://openalex.org/C149782125 |
| concepts[11].level | 1 |
| concepts[11].score | 0.4058927297592163 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q160039 |
| concepts[11].display_name | Econometrics |
| concepts[12].id | https://openalex.org/C160234255 |
| concepts[12].level | 3 |
| concepts[12].score | 0.33665746450424194 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q812535 |
| concepts[12].display_name | Bayesian inference |
| concepts[13].id | https://openalex.org/C33923547 |
| concepts[13].level | 0 |
| concepts[13].score | 0.2725215256214142 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[13].display_name | Mathematics |
| concepts[14].id | https://openalex.org/C60644358 |
| concepts[14].level | 1 |
| concepts[14].score | 0.20048293471336365 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[14].display_name | Bioinformatics |
| keywords[0].id | https://openalex.org/keywords/overdispersion |
| keywords[0].score | 0.7616919279098511 |
| keywords[0].display_name | Overdispersion |
| keywords[1].id | https://openalex.org/keywords/negative-binomial-distribution |
| keywords[1].score | 0.7409213781356812 |
| keywords[1].display_name | Negative binomial distribution |
| keywords[2].id | https://openalex.org/keywords/count-data |
| keywords[2].score | 0.584478497505188 |
| keywords[2].display_name | Count data |
| keywords[3].id | https://openalex.org/keywords/microbiome |
| keywords[3].score | 0.5826258063316345 |
| keywords[3].display_name | Microbiome |
| keywords[4].id | https://openalex.org/keywords/covariate |
| keywords[4].score | 0.5821583867073059 |
| keywords[4].display_name | Covariate |
| keywords[5].id | https://openalex.org/keywords/statistics |
| keywords[5].score | 0.5059203505516052 |
| keywords[5].display_name | Statistics |
| keywords[6].id | https://openalex.org/keywords/bayes-factor |
| keywords[6].score | 0.49013885855674744 |
| keywords[6].display_name | Bayes factor |
| keywords[7].id | https://openalex.org/keywords/biology |
| keywords[7].score | 0.46630609035491943 |
| keywords[7].display_name | Biology |
| keywords[8].id | https://openalex.org/keywords/regression |
| keywords[8].score | 0.46408554911613464 |
| keywords[8].display_name | Regression |
| keywords[9].id | https://openalex.org/keywords/poisson-distribution |
| keywords[9].score | 0.4227152466773987 |
| keywords[9].display_name | Poisson distribution |
| keywords[10].id | https://openalex.org/keywords/bayesian-probability |
| keywords[10].score | 0.4134995937347412 |
| keywords[10].display_name | Bayesian probability |
| keywords[11].id | https://openalex.org/keywords/econometrics |
| keywords[11].score | 0.4058927297592163 |
| keywords[11].display_name | Econometrics |
| keywords[12].id | https://openalex.org/keywords/bayesian-inference |
| keywords[12].score | 0.33665746450424194 |
| keywords[12].display_name | Bayesian inference |
| keywords[13].id | https://openalex.org/keywords/mathematics |
| keywords[13].score | 0.2725215256214142 |
| keywords[13].display_name | Mathematics |
| keywords[14].id | https://openalex.org/keywords/bioinformatics |
| keywords[14].score | 0.20048293471336365 |
| keywords[14].display_name | Bioinformatics |
| language | en |
| locations[0].id | doi:10.1128/msystems.01345-24 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210181942 |
| locations[0].source.issn | 2379-5077 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2379-5077 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | mSystems |
| locations[0].source.host_organization | https://openalex.org/P4310320263 |
| locations[0].source.host_organization_name | American Society for Microbiology |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320263 |
| locations[0].source.host_organization_lineage_names | American Society for Microbiology |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | mSystems |
| locations[0].landing_page_url | https://doi.org/10.1128/msystems.01345-24 |
| locations[1].id | pmid:39570024 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | mSystems |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39570024 |
| locations[2].id | pmh:oai:doaj.org/article:e0768c73ebaa4d7083e39b7b76cb6513 |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | mSystems, Vol 9, Iss 12 (2024) |
| locations[2].landing_page_url | https://doaj.org/article/e0768c73ebaa4d7083e39b7b76cb6513 |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:11651110 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | mSystems |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11651110 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5108949548 |
| authorships[0].author.orcid | https://orcid.org/0009-0001-5085-8422 |
| authorships[0].author.display_name | Jian Huang |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I102208913 |
| authorships[0].affiliations[0].raw_affiliation_string | School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I102208913 |
| authorships[0].affiliations[1].raw_affiliation_string | Key Laboratory of Xinjiang Special Probiotics and Dairy Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[0].institutions[0].id | https://openalex.org/I102208913 |
| authorships[0].institutions[0].ror | https://ror.org/04x0kvm78 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I102208913 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Shihezi University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jian Huang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Key Laboratory of Xinjiang Special Probiotics and Dairy Technology, Shihezi University, Shihezi, Xinjiang, China, School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[1].author.id | https://openalex.org/A5108321575 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Yanzhuan Lu |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I102208913 |
| authorships[1].affiliations[0].raw_affiliation_string | Key Laboratory of Xinjiang Special Probiotics and Dairy Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I102208913 |
| authorships[1].affiliations[1].raw_affiliation_string | School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[1].institutions[0].id | https://openalex.org/I102208913 |
| authorships[1].institutions[0].ror | https://ror.org/04x0kvm78 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I102208913 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Shihezi University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Yanzhuan Lu |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Key Laboratory of Xinjiang Special Probiotics and Dairy Technology, Shihezi University, Shihezi, Xinjiang, China, School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[2].author.id | https://openalex.org/A5069260620 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-8511-4546 |
| authorships[2].author.display_name | Fengwei Tian |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I111599522, https://openalex.org/I4210152154 |
| authorships[2].affiliations[0].raw_affiliation_string | State Key Laboratory of Food Science and Resources, Jiangnan University, Wuxi, Jiangsu, China |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I111599522 |
| authorships[2].affiliations[1].raw_affiliation_string | School of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu, China |
| authorships[2].institutions[0].id | https://openalex.org/I111599522 |
| authorships[2].institutions[0].ror | https://ror.org/04mkzax54 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I111599522 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Jiangnan University |
| authorships[2].institutions[1].id | https://openalex.org/I4210152154 |
| authorships[2].institutions[1].ror | https://ror.org/04zgj7914 |
| authorships[2].institutions[1].type | facility |
| authorships[2].institutions[1].lineage | https://openalex.org/I111599522, https://openalex.org/I141649914, https://openalex.org/I4210152154 |
| authorships[2].institutions[1].country_code | CN |
| authorships[2].institutions[1].display_name | State Key Laboratory of Food Science and Technology |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Fengwei Tian |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | School of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu, China, State Key Laboratory of Food Science and Resources, Jiangnan University, Wuxi, Jiangsu, China |
| authorships[3].author.id | https://openalex.org/A5081441019 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-4876-589X |
| authorships[3].author.display_name | Yongqing Ni |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I102208913 |
| authorships[3].affiliations[0].raw_affiliation_string | School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I102208913 |
| authorships[3].affiliations[1].raw_affiliation_string | Key Laboratory of Xinjiang Special Probiotics and Dairy Technology, Shihezi University, Shihezi, Xinjiang, China |
| authorships[3].institutions[0].id | https://openalex.org/I102208913 |
| authorships[3].institutions[0].ror | https://ror.org/04x0kvm78 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I102208913 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Shihezi University |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Yongqing Ni |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Key Laboratory of Xinjiang Special Probiotics and Dairy Technology, Shihezi University, Shihezi, Xinjiang, China, School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1128/msystems.01345-24 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Association of body index with fecal microbiome in children cohorts with ethnic–geographic factor interaction: accurately using a Bayesian zero-inflated negative binomial regression model |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-23T05:10:03.516525 |
| primary_topic.id | https://openalex.org/T10066 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Gut microbiota and health |
| related_works | https://openalex.org/W2189104843, https://openalex.org/W2885569989, https://openalex.org/W3162047386, https://openalex.org/W2556931687, https://openalex.org/W3134768068, https://openalex.org/W3122903216, https://openalex.org/W2041256494, https://openalex.org/W1967330102, https://openalex.org/W2991740003, https://openalex.org/W2047828157 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 4 |
| best_oa_location.id | doi:10.1128/msystems.01345-24 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210181942 |
| best_oa_location.source.issn | 2379-5077 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2379-5077 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | mSystems |
| best_oa_location.source.host_organization | https://openalex.org/P4310320263 |
| best_oa_location.source.host_organization_name | American Society for Microbiology |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320263 |
| best_oa_location.source.host_organization_lineage_names | American Society for Microbiology |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | mSystems |
| best_oa_location.landing_page_url | https://doi.org/10.1128/msystems.01345-24 |
| primary_location.id | doi:10.1128/msystems.01345-24 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210181942 |
| primary_location.source.issn | 2379-5077 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2379-5077 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | mSystems |
| primary_location.source.host_organization | https://openalex.org/P4310320263 |
| primary_location.source.host_organization_name | American Society for Microbiology |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320263 |
| primary_location.source.host_organization_lineage_names | American Society for Microbiology |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | mSystems |
| primary_location.landing_page_url | https://doi.org/10.1128/msystems.01345-24 |
| publication_date | 2024-11-21 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2162088497, https://openalex.org/W2264701027, https://openalex.org/W2913097383, https://openalex.org/W1516274582, https://openalex.org/W2395461901, https://openalex.org/W2890189910, https://openalex.org/W4236036775, https://openalex.org/W3100759574, https://openalex.org/W3097964496, https://openalex.org/W1981457167, https://openalex.org/W2255524332, https://openalex.org/W3173323489, https://openalex.org/W3154090598, https://openalex.org/W2312269541, https://openalex.org/W4243621906, https://openalex.org/W4223964666, https://openalex.org/W2955435388, https://openalex.org/W3003072093, https://openalex.org/W2894062910, https://openalex.org/W3198549346, https://openalex.org/W4292296130, https://openalex.org/W3184541229, https://openalex.org/W3196032280, https://openalex.org/W3108994080, https://openalex.org/W3091376159, https://openalex.org/W2051800985, https://openalex.org/W2119634512, https://openalex.org/W2054253051, https://openalex.org/W2476077119, https://openalex.org/W6979014210, https://openalex.org/W4292528167, https://openalex.org/W346827628, https://openalex.org/W2124351063, https://openalex.org/W2518795824, https://openalex.org/W2034285706, https://openalex.org/W4399545607, https://openalex.org/W30139279, https://openalex.org/W1747046542, https://openalex.org/W4386224032, https://openalex.org/W4306412058, https://openalex.org/W1974858315, https://openalex.org/W1581231885, https://openalex.org/W4320913259, https://openalex.org/W2901939432, https://openalex.org/W3030238839, https://openalex.org/W2587304885, https://openalex.org/W2584763199, https://openalex.org/W3092986592, https://openalex.org/W2803790829, https://openalex.org/W4317666413, https://openalex.org/W3137678236, https://openalex.org/W2936328687, https://openalex.org/W2115742500, https://openalex.org/W4200567941, https://openalex.org/W2910147931, https://openalex.org/W2901965455, https://openalex.org/W3080954194, https://openalex.org/W2615884100, https://openalex.org/W2886557383, https://openalex.org/W2891673573, https://openalex.org/W2994811958, https://openalex.org/W2088815801, https://openalex.org/W3211779138, https://openalex.org/W3171259862, https://openalex.org/W2102892532, https://openalex.org/W2473355215, https://openalex.org/W3007945801, https://openalex.org/W2953583829, https://openalex.org/W2756958651, https://openalex.org/W2165421558 |
| referenced_works_count | 70 |
| abstract_inverted_index.a | 35, 154, 280 |
| abstract_inverted_index.10 | 52 |
| abstract_inverted_index.In | 186 |
| abstract_inverted_index.We | 295 |
| abstract_inverted_index.an | 16, 62, 297 |
| abstract_inverted_index.as | 37, 131, 160, 235, 245, 267 |
| abstract_inverted_index.be | 212, 243, 288 |
| abstract_inverted_index.by | 50, 110 |
| abstract_inverted_index.in | 107, 145, 164, 178, 195, 254, 291, 344 |
| abstract_inverted_index.is | 72, 201, 209, 225, 265, 303, 329, 350 |
| abstract_inverted_index.of | 4, 25, 74, 88, 124, 134, 156, 174, 190, 218, 231, 247, 262, 270, 282, 305, 319, 339 |
| abstract_inverted_index.on | 9, 90, 321 |
| abstract_inverted_index.to | 19, 46, 142, 211, 214, 260, 331 |
| abstract_inverted_index.we | 60, 115, 150 |
| abstract_inverted_index.Our | 93, 337 |
| abstract_inverted_index.The | 1, 223 |
| abstract_inverted_index.age | 238 |
| abstract_inverted_index.and | 44, 84, 103, 112, 172, 198, 203, 239, 315, 324, 334, 347, 352 |
| abstract_inverted_index.are | 40, 257 |
| abstract_inverted_index.can | 242, 287 |
| abstract_inverted_index.for | 56 |
| abstract_inverted_index.its | 327 |
| abstract_inverted_index.our | 140, 188, 207 |
| abstract_inverted_index.the | 10, 23, 86, 122, 132, 169, 215, 232, 255, 317 |
| abstract_inverted_index.BMI, | 240 |
| abstract_inverted_index.INLA | 104, 335 |
| abstract_inverted_index.ZINB | 299 |
| abstract_inverted_index.also | 33 |
| abstract_inverted_index.data | 8, 39, 144, 177, 197, 346 |
| abstract_inverted_index.host | 28, 82, 313 |
| abstract_inverted_index.into | 22 |
| abstract_inverted_index.most | 179 |
| abstract_inverted_index.only | 153 |
| abstract_inverted_index.real | 146, 165 |
| abstract_inverted_index.such | 234 |
| abstract_inverted_index.taxa | 78, 157, 253, 309 |
| abstract_inverted_index.that | 71, 117, 152, 168, 302, 326 |
| abstract_inverted_index.this | 31 |
| abstract_inverted_index.type | 281 |
| abstract_inverted_index.used | 244 |
| abstract_inverted_index.were | 158, 183 |
| abstract_inverted_index.with | 15, 27, 80, 100, 127, 228, 311 |
| abstract_inverted_index.zero | 47, 162 |
| abstract_inverted_index.(HTS) | 7 |
| abstract_inverted_index.Taken | 205 |
| abstract_inverted_index.body, | 233 |
| abstract_inverted_index.child | 272 |
| abstract_inverted_index.count | 176 |
| abstract_inverted_index.data, | 59, 166 |
| abstract_inverted_index.delve | 20 |
| abstract_inverted_index.found | 151 |
| abstract_inverted_index.human | 248 |
| abstract_inverted_index.model | 70, 95, 125, 141, 301 |
| abstract_inverted_index.poses | 34 |
| abstract_inverted_index.prone | 45 |
| abstract_inverted_index.taxa, | 323 |
| abstract_inverted_index.taxa. | 92 |
| abstract_inverted_index.these | 91, 322 |
| abstract_inverted_index.which | 241, 252 |
| abstract_inverted_index.(ZINB) | 68, 278 |
| abstract_inverted_index.Hurdle | 102, 333 |
| abstract_inverted_index.affect | 121 |
| abstract_inverted_index.deeper | 21 |
| abstract_inverted_index.growth | 3, 32, 273 |
| abstract_inverted_index.having | 161 |
| abstract_inverted_index.linear | 285 |
| abstract_inverted_index.method | 208 |
| abstract_inverted_index.model, | 279, 286 |
| abstract_inverted_index.models | 55 |
| abstract_inverted_index.number | 133 |
| abstract_inverted_index.overly | 184 |
| abstract_inverted_index.subset | 155 |
| abstract_inverted_index.Herein, | 49 |
| abstract_inverted_index.capable | 73, 304 |
| abstract_inverted_index.closely | 226, 258 |
| abstract_inverted_index.cohort, | 149 |
| abstract_inverted_index.complex | 292 |
| abstract_inverted_index.confirm | 116 |
| abstract_inverted_index.defects | 128 |
| abstract_inverted_index.effects | 87, 318 |
| abstract_inverted_index.health. | 249 |
| abstract_inverted_index.height, | 236 |
| abstract_inverted_index.markers | 269 |
| abstract_inverted_index.modeled | 290 |
| abstract_inverted_index.models, | 105 |
| abstract_inverted_index.models. | 336 |
| abstract_inverted_index.present | 296 |
| abstract_inverted_index.related | 259 |
| abstract_inverted_index.samples | 136 |
| abstract_inverted_index.sparse, | 42 |
| abstract_inverted_index.studies | 182, 217 |
| abstract_inverted_index.various | 219 |
| abstract_inverted_index.weight, | 237 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.Bayesian | 64, 283 |
| abstract_inverted_index.However, | 30 |
| abstract_inverted_index.abundant | 77, 308 |
| abstract_inverted_index.accuracy | 98, 123, 328 |
| abstract_inverted_index.amplicon | 143 |
| abstract_inverted_index.analyzed | 135 |
| abstract_inverted_index.applying | 139 |
| abstract_inverted_index.binomial | 67, 277 |
| abstract_inverted_index.children | 148 |
| abstract_inverted_index.compared | 99 |
| abstract_inverted_index.complex, | 41 |
| abstract_inverted_index.counting | 54 |
| abstract_inverted_index.distinct | 53 |
| abstract_inverted_index.exhibits | 96 |
| abstract_inverted_index.expected | 210 |
| abstract_inverted_index.extended | 213 |
| abstract_inverted_index.measures | 246 |
| abstract_inverted_index.negative | 66, 276 |
| abstract_inverted_index.physical | 229, 263 |
| abstract_inverted_index.pipeline | 189, 338 |
| abstract_inverted_index.presents | 13 |
| abstract_inverted_index.previous | 180 |
| abstract_inverted_index.proposed | 61, 94 |
| abstract_inverted_index.regional | 271 |
| abstract_inverted_index.relevant | 199, 348 |
| abstract_inverted_index.results, | 126 |
| abstract_inverted_index.superior | 330 |
| abstract_inverted_index.systems. | 294 |
| abstract_inverted_index.valuable | 266 |
| abstract_inverted_index.Moreover, | 114 |
| abstract_inverted_index.abundance | 194, 343 |
| abstract_inverted_index.analyzing | 57 |
| abstract_inverted_index.bacterial | 192, 341 |
| abstract_inverted_index.discrete, | 43 |
| abstract_inverted_index.dogmatic. | 185 |
| abstract_inverted_index.effective | 202, 351 |
| abstract_inverted_index.excellent | 97 |
| abstract_inverted_index.feasible. | 204, 353 |
| abstract_inverted_index.gradually | 129 |
| abstract_inverted_index.inflation | 111, 163 |
| abstract_inverted_index.microbial | 11, 38, 175, 268 |
| abstract_inverted_index.practice, | 187 |
| abstract_inverted_index.scenarios | 108 |
| abstract_inverted_index.simulated | 58 |
| abstract_inverted_index.together, | 206 |
| abstract_inverted_index.utilizing | 51 |
| abstract_inverted_index.Accurately | 250 |
| abstract_inverted_index.IMPORTANCE | 222 |
| abstract_inverted_index.associated | 79, 227, 310 |
| abstract_inverted_index.biological | 293 |
| abstract_inverted_index.challenge, | 36 |
| abstract_inverted_index.covariates | 89, 200, 320, 349 |
| abstract_inverted_index.dispersion | 118 |
| abstract_inverted_index.especially | 106 |
| abstract_inverted_index.identified | 159 |
| abstract_inverted_index.increases. | 137 |
| abstract_inverted_index.indicators | 230, 261 |
| abstract_inverted_index.inflation. | 48 |
| abstract_inverted_index.innovative | 63, 298 |
| abstract_inverted_index.microbiome | 181, 196, 224, 256, 345 |
| abstract_inverted_index.microbiota | 216 |
| abstract_inverted_index.parameters | 119 |
| abstract_inverted_index.phenotype. | 29 |
| abstract_inverted_index.phenotypes | 83, 314 |
| abstract_inverted_index.prevailing | 170 |
| abstract_inverted_index.processing | 173 |
| abstract_inverted_index.regression | 69, 300 |
| abstract_inverted_index.sequencing | 6 |
| abstract_inverted_index.suggesting | 167 |
| abstract_inverted_index.alleviating | 130 |
| abstract_inverted_index.association | 24 |
| abstract_inverted_index.communities | 12 |
| abstract_inverted_index.demonstrate | 325 |
| abstract_inverted_index.development | 264 |
| abstract_inverted_index.distinctive | 81, 312 |
| abstract_inverted_index.effectively | 289 |
| abstract_inverted_index.exponential | 2 |
| abstract_inverted_index.generalized | 284 |
| abstract_inverted_index.identifying | 75, 251, 306 |
| abstract_inverted_index.integrating | 191, 340 |
| abstract_inverted_index.opportunity | 18 |
| abstract_inverted_index.quantifying | 85, 316 |
| abstract_inverted_index.researchers | 14 |
| abstract_inverted_index.traditional | 332 |
| abstract_inverted_index.trajectory. | 274 |
| abstract_inverted_index.Subsequently | 138 |
| abstract_inverted_index.conventional | 101 |
| abstract_inverted_index.differential | 193, 342 |
| abstract_inverted_index.multi-cohort | 220 |
| abstract_inverted_index.multi-ethnic | 147 |
| abstract_inverted_index.populations. | 221 |
| abstract_inverted_index.unparalleled | 17 |
| abstract_inverted_index.Zero-inflated | 275 |
| abstract_inverted_index.characterized | 109 |
| abstract_inverted_index.significantly | 120 |
| abstract_inverted_index.understanding | 171 |
| abstract_inverted_index.zero-inflated | 65 |
| abstract_inverted_index.differentially | 76, 307 |
| abstract_inverted_index.microorganisms | 26 |
| abstract_inverted_index.high-throughput | 5 |
| abstract_inverted_index.overdispersion. | 113 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 4 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/8 |
| sustainable_development_goals[0].score | 0.5400000214576721 |
| sustainable_development_goals[0].display_name | Decent work and economic growth |
| citation_normalized_percentile.value | 0.63648822 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |