Atomistic Polymer Modeling: Recent Advances and Challenges in Building and Parametrization Workflows Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1021/acs.macromol.5c01166
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1021/acs.macromol.5c01166
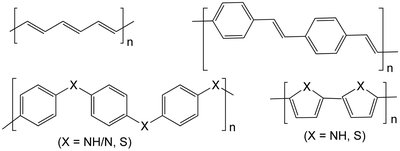
Synthetic polymers are a broad and versatile class of soft materials covering a large chemical space. “Computational microscopy” approaches such as atomistic molecular dynamics (MD) simulations are an effective tool to validate and rationalize experimental data for structure–property characterization. The predictive quality of MD simulations and the properties derived from them are primarily driven by the accuracy and relevance of the force field used to represent the system. While biomolecular simulation (nucleic acids, proteins) workflows benefit from dedicated toolkits and domain-specific force fields, the modeling of synthetic polymers has not progressed to the same extent. This perspective will discuss recent efforts to improve system building and parametrization workflows for synthetic polymers, and the unique challenges differentiating them from biopolymers. We will outline shortcomings in established workflows, review best practices for FAIR polymer simulations, and highlight new tools/workflows leveraging cheminformatics, direct chemical perception, and neural networks.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1021/acs.macromol.5c01166
- OA Status
- hybrid
- References
- 166
- OpenAlex ID
- https://openalex.org/W4415615509
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4415615509Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1021/acs.macromol.5c01166Digital Object Identifier
- Title
-
Atomistic Polymer Modeling: Recent Advances and Challenges in Building and Parametrization WorkflowsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-10-28Full publication date if available
- Authors
-
Hannah N. Turney, Micaela MattaList of authors in order
- Landing page
-
https://doi.org/10.1021/acs.macromol.5c01166Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1021/acs.macromol.5c01166Direct OA link when available
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
166Number of works referenced by this work
Full payload
| id | https://openalex.org/W4415615509 |
|---|---|
| doi | https://doi.org/10.1021/acs.macromol.5c01166 |
| ids.doi | https://doi.org/10.1021/acs.macromol.5c01166 |
| ids.openalex | https://openalex.org/W4415615509 |
| fwci | |
| type | article |
| title | Atomistic Polymer Modeling: Recent Advances and Challenges in Building and Parametrization Workflows |
| biblio.issue | 21 |
| biblio.volume | 58 |
| biblio.last_page | 11522 |
| biblio.first_page | 11509 |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| language | en |
| locations[0].id | doi:10.1021/acs.macromol.5c01166 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S177196338 |
| locations[0].source.issn | 0024-9297, 1520-5835 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0024-9297 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Macromolecules |
| locations[0].source.host_organization | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_name | American Chemical Society |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320006 |
| locations[0].source.host_organization_lineage_names | American Chemical Society |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Macromolecules |
| locations[0].landing_page_url | https://doi.org/10.1021/acs.macromol.5c01166 |
| locations[1].id | pmh:oai:europepmc.org:11422116 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400806 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | Europe PMC (PubMed Central) |
| locations[1].source.host_organization | https://openalex.org/I1303153112 |
| locations[1].source.host_organization_name | European Bioinformatics Institute |
| locations[1].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[1].license | cc-by |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Text |
| locations[1].license_id | https://openalex.org/licenses/cc-by |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/12613811 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5081435162 |
| authorships[0].author.orcid | https://orcid.org/0009-0002-3298-0309 |
| authorships[0].author.display_name | Hannah N. Turney |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I183935753 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Chemistry, King's College London Strand Campus (East Wing), London WC2R 2LS, United Kingdom |
| authorships[0].institutions[0].id | https://openalex.org/I183935753 |
| authorships[0].institutions[0].ror | https://ror.org/0220mzb33 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I124357947, https://openalex.org/I183935753 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | King's College London |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Hannah N. Turney |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Chemistry, King's College London Strand Campus (East Wing), London WC2R 2LS, United Kingdom |
| authorships[1].author.id | https://openalex.org/A5069673994 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-9852-3154 |
| authorships[1].author.display_name | Micaela Matta |
| authorships[1].countries | GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I183935753 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Chemistry, King's College London Strand Campus (East Wing), London WC2R 2LS, United Kingdom |
| authorships[1].institutions[0].id | https://openalex.org/I183935753 |
| authorships[1].institutions[0].ror | https://ror.org/0220mzb33 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I124357947, https://openalex.org/I183935753 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | King's College London |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | Micaela Matta |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Department of Chemistry, King's College London Strand Campus (East Wing), London WC2R 2LS, United Kingdom |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1021/acs.macromol.5c01166 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-28T00:00:00 |
| display_name | Atomistic Polymer Modeling: Recent Advances and Challenges in Building and Parametrization Workflows |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-12T23:11:45.498971 |
| primary_topic | |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1021/acs.macromol.5c01166 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S177196338 |
| best_oa_location.source.issn | 0024-9297, 1520-5835 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0024-9297 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Macromolecules |
| best_oa_location.source.host_organization | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_name | American Chemical Society |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| best_oa_location.source.host_organization_lineage_names | American Chemical Society |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Macromolecules |
| best_oa_location.landing_page_url | https://doi.org/10.1021/acs.macromol.5c01166 |
| primary_location.id | doi:10.1021/acs.macromol.5c01166 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S177196338 |
| primary_location.source.issn | 0024-9297, 1520-5835 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0024-9297 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Macromolecules |
| primary_location.source.host_organization | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_name | American Chemical Society |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320006 |
| primary_location.source.host_organization_lineage_names | American Chemical Society |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Macromolecules |
| primary_location.landing_page_url | https://doi.org/10.1021/acs.macromol.5c01166 |
| publication_date | 2025-10-28 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W4409148148, https://openalex.org/W4402063381, https://openalex.org/W4317104064, https://openalex.org/W4389157189, https://openalex.org/W2888400067, https://openalex.org/W4379012421, https://openalex.org/W4210600092, https://openalex.org/W4311205118, https://openalex.org/W2087032263, https://openalex.org/W2641076635, https://openalex.org/W3190984626, https://openalex.org/W3128633116, https://openalex.org/W4288067320, https://openalex.org/W4388735746, https://openalex.org/W2510236435, https://openalex.org/W4289207127, https://openalex.org/W4408043087, https://openalex.org/W2946873322, https://openalex.org/W3113492468, https://openalex.org/W2805462747, https://openalex.org/W2569644332, https://openalex.org/W2395345938, https://openalex.org/W2969961665, https://openalex.org/W2963426345, https://openalex.org/W4398143808, https://openalex.org/W4409620975, https://openalex.org/W2948312333, https://openalex.org/W4220928420, https://openalex.org/W4410166836, https://openalex.org/W4205573868, https://openalex.org/W2912807874, https://openalex.org/W2302501749, https://openalex.org/W3015598776, https://openalex.org/W4392498998, https://openalex.org/W4225262991, https://openalex.org/W3206968565, https://openalex.org/W4385519663, https://openalex.org/W2136815817, https://openalex.org/W4362523109, https://openalex.org/W4321435541, https://openalex.org/W4213034697, https://openalex.org/W2052563605, https://openalex.org/W4282823776, https://openalex.org/W4405306979, https://openalex.org/W4396835380, https://openalex.org/W4387440676, https://openalex.org/W4308943751, https://openalex.org/W2511309442, https://openalex.org/W4411715505, https://openalex.org/W2946410014, https://openalex.org/W3201732108, https://openalex.org/W4317215121, https://openalex.org/W3096386116, https://openalex.org/W2973074478, https://openalex.org/W4383955629, https://openalex.org/W3162823304, https://openalex.org/W4408785841, https://openalex.org/W6940671333, https://openalex.org/W2943206967, https://openalex.org/W2898210859, https://openalex.org/W3177828909, https://openalex.org/W2031168104, https://openalex.org/W2949223833, https://openalex.org/W2909063104, https://openalex.org/W2078374001, https://openalex.org/W1976161355, https://openalex.org/W4402205029, https://openalex.org/W2003949305, https://openalex.org/W4220855002, https://openalex.org/W4308586705, https://openalex.org/W2540491215, https://openalex.org/W2510475640, https://openalex.org/W2147988069, https://openalex.org/W6967424702, https://openalex.org/W4303579435, https://openalex.org/W2154449781, https://openalex.org/W4379768996, https://openalex.org/W3145808230, https://openalex.org/W2472797552, https://openalex.org/W4408514545, https://openalex.org/W3128698945, https://openalex.org/W1992339286, https://openalex.org/W2565058004, https://openalex.org/W4313251368, https://openalex.org/W2160330738, https://openalex.org/W2062618849, https://openalex.org/W4311882199, https://openalex.org/W3009719057, https://openalex.org/W2932067160, https://openalex.org/W2608747371, https://openalex.org/W4402762870, https://openalex.org/W4404237118, https://openalex.org/W4392979718, https://openalex.org/W2060433756, https://openalex.org/W2963268124, https://openalex.org/W2921209122, https://openalex.org/W2037788435, https://openalex.org/W2606142660, https://openalex.org/W2898021236, https://openalex.org/W2041189078, https://openalex.org/W2072683434, https://openalex.org/W4391476245, https://openalex.org/W2095145098, https://openalex.org/W2122199548, https://openalex.org/W4319294969, https://openalex.org/W4403595529, https://openalex.org/W3208561583, https://openalex.org/W3123030707, https://openalex.org/W4281391354, https://openalex.org/W2072976471, https://openalex.org/W3043735243, https://openalex.org/W2091654157, https://openalex.org/W4404645058, https://openalex.org/W2937468399, https://openalex.org/W2885431651, https://openalex.org/W4206277361, https://openalex.org/W2321111305, https://openalex.org/W1726671544, https://openalex.org/W2969829863, https://openalex.org/W3082649560, https://openalex.org/W4404125054, https://openalex.org/W2471657669, https://openalex.org/W3037545945, https://openalex.org/W2317941165, https://openalex.org/W2039588355, https://openalex.org/W3136779761, https://openalex.org/W4376226284, https://openalex.org/W4406219082, https://openalex.org/W4295096853, https://openalex.org/W4396733928, https://openalex.org/W6891811738, https://openalex.org/W3133931590, https://openalex.org/W4283021369, https://openalex.org/W2547447472, https://openalex.org/W4406385288, https://openalex.org/W3163857962, https://openalex.org/W4387522931, https://openalex.org/W4386436455, https://openalex.org/W4406154662, https://openalex.org/W4386954398, https://openalex.org/W4410996149, https://openalex.org/W4411424580, https://openalex.org/W4308653706, https://openalex.org/W2111917512, https://openalex.org/W2150789776, https://openalex.org/W3200391459, https://openalex.org/W4315606103, https://openalex.org/W4407783312, https://openalex.org/W2096769282, https://openalex.org/W4399808054, https://openalex.org/W6913074012, https://openalex.org/W4388455891, https://openalex.org/W4411471785, https://openalex.org/W4387655061, https://openalex.org/W4412008160, https://openalex.org/W6929166631, https://openalex.org/W2754122850, https://openalex.org/W2003404863, https://openalex.org/W4407108797, https://openalex.org/W3048327694, https://openalex.org/W4367836627, https://openalex.org/W4386283721, https://openalex.org/W4388638767, https://openalex.org/W4297914802, https://openalex.org/W4321367323, https://openalex.org/W4407645172 |
| referenced_works_count | 166 |
| abstract_inverted_index.a | 3, 12 |
| abstract_inverted_index.MD | 43 |
| abstract_inverted_index.We | 119 |
| abstract_inverted_index.an | 27 |
| abstract_inverted_index.as | 20 |
| abstract_inverted_index.by | 54 |
| abstract_inverted_index.in | 123 |
| abstract_inverted_index.of | 8, 42, 59, 85 |
| abstract_inverted_index.to | 30, 64, 91, 101 |
| abstract_inverted_index.The | 39 |
| abstract_inverted_index.and | 5, 32, 45, 57, 79, 105, 111, 133, 142 |
| abstract_inverted_index.are | 2, 26, 51 |
| abstract_inverted_index.for | 36, 108, 129 |
| abstract_inverted_index.has | 88 |
| abstract_inverted_index.new | 135 |
| abstract_inverted_index.not | 89 |
| abstract_inverted_index.the | 46, 55, 60, 66, 83, 92, 112 |
| abstract_inverted_index.(MD) | 24 |
| abstract_inverted_index.FAIR | 130 |
| abstract_inverted_index.This | 95 |
| abstract_inverted_index.best | 127 |
| abstract_inverted_index.data | 35 |
| abstract_inverted_index.from | 49, 76, 117 |
| abstract_inverted_index.same | 93 |
| abstract_inverted_index.soft | 9 |
| abstract_inverted_index.such | 19 |
| abstract_inverted_index.them | 50, 116 |
| abstract_inverted_index.tool | 29 |
| abstract_inverted_index.used | 63 |
| abstract_inverted_index.will | 97, 120 |
| abstract_inverted_index.While | 68 |
| abstract_inverted_index.broad | 4 |
| abstract_inverted_index.class | 7 |
| abstract_inverted_index.field | 62 |
| abstract_inverted_index.force | 61, 81 |
| abstract_inverted_index.large | 13 |
| abstract_inverted_index.acids, | 72 |
| abstract_inverted_index.direct | 139 |
| abstract_inverted_index.driven | 53 |
| abstract_inverted_index.neural | 143 |
| abstract_inverted_index.recent | 99 |
| abstract_inverted_index.review | 126 |
| abstract_inverted_index.space. | 15 |
| abstract_inverted_index.system | 103 |
| abstract_inverted_index.unique | 113 |
| abstract_inverted_index.benefit | 75 |
| abstract_inverted_index.derived | 48 |
| abstract_inverted_index.discuss | 98 |
| abstract_inverted_index.efforts | 100 |
| abstract_inverted_index.extent. | 94 |
| abstract_inverted_index.fields, | 82 |
| abstract_inverted_index.improve | 102 |
| abstract_inverted_index.outline | 121 |
| abstract_inverted_index.polymer | 131 |
| abstract_inverted_index.quality | 41 |
| abstract_inverted_index.system. | 67 |
| abstract_inverted_index.(nucleic | 71 |
| abstract_inverted_index.accuracy | 56 |
| abstract_inverted_index.building | 104 |
| abstract_inverted_index.chemical | 14, 140 |
| abstract_inverted_index.covering | 11 |
| abstract_inverted_index.dynamics | 23 |
| abstract_inverted_index.modeling | 84 |
| abstract_inverted_index.polymers | 1, 87 |
| abstract_inverted_index.toolkits | 78 |
| abstract_inverted_index.validate | 31 |
| abstract_inverted_index.Synthetic | 0 |
| abstract_inverted_index.atomistic | 21 |
| abstract_inverted_index.dedicated | 77 |
| abstract_inverted_index.effective | 28 |
| abstract_inverted_index.highlight | 134 |
| abstract_inverted_index.materials | 10 |
| abstract_inverted_index.molecular | 22 |
| abstract_inverted_index.networks. | 144 |
| abstract_inverted_index.polymers, | 110 |
| abstract_inverted_index.practices | 128 |
| abstract_inverted_index.primarily | 52 |
| abstract_inverted_index.proteins) | 73 |
| abstract_inverted_index.relevance | 58 |
| abstract_inverted_index.represent | 65 |
| abstract_inverted_index.synthetic | 86, 109 |
| abstract_inverted_index.versatile | 6 |
| abstract_inverted_index.workflows | 74, 107 |
| abstract_inverted_index.approaches | 18 |
| abstract_inverted_index.challenges | 114 |
| abstract_inverted_index.leveraging | 137 |
| abstract_inverted_index.predictive | 40 |
| abstract_inverted_index.progressed | 90 |
| abstract_inverted_index.properties | 47 |
| abstract_inverted_index.simulation | 70 |
| abstract_inverted_index.workflows, | 125 |
| abstract_inverted_index.established | 124 |
| abstract_inverted_index.perception, | 141 |
| abstract_inverted_index.perspective | 96 |
| abstract_inverted_index.rationalize | 33 |
| abstract_inverted_index.simulations | 25, 44 |
| abstract_inverted_index.biomolecular | 69 |
| abstract_inverted_index.biopolymers. | 118 |
| abstract_inverted_index.experimental | 34 |
| abstract_inverted_index.shortcomings | 122 |
| abstract_inverted_index.simulations, | 132 |
| abstract_inverted_index.microscopy” | 17 |
| abstract_inverted_index.differentiating | 115 |
| abstract_inverted_index.domain-specific | 80 |
| abstract_inverted_index.parametrization | 106 |
| abstract_inverted_index.tools/workflows | 136 |
| abstract_inverted_index.cheminformatics, | 138 |
| abstract_inverted_index.“Computational | 16 |
| abstract_inverted_index.characterization. | 38 |
| abstract_inverted_index.structure–property | 37 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5069673994 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 2 |
| corresponding_institution_ids | https://openalex.org/I183935753 |
| citation_normalized_percentile |