AttnTAP: A Dual-input Framework Incorporating the Attention Mechanism for Accurately Predicting TCR-peptide Binding Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.3389/fgene.2022.942491
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.3389/fgene.2022.942491
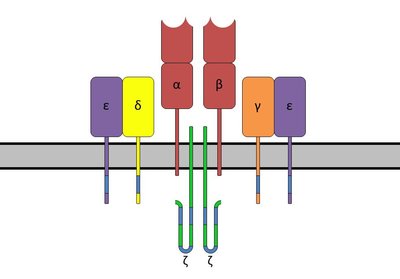
T-cell receptors (TCRs) are formed by random recombination of genomic precursor elements, some of which mediate the recognition of cancer-associated antigens. Due to the complicated process of T-cell immune response and limited biological empirical evidence, the practical strategy for identifying TCRs and their recognized peptides is the computational prediction from population and/or individual TCR repertoires. In recent years, several machine/deep learning-based approaches have been proposed for TCR-peptide binding prediction. However, the predictive performances of these methods can be further improved by overcoming several significant flaws in neural network design. The interrelationship between amino acids in TCRs is critical for TCR antigen recognition, which was not properly considered by the existing methods. They also did not pay more attention to the amino acids that play a significant role in antigen-binding specificity. Moreover, complex networks tended to increase the risk of overfitting and computational costs. In this study, we developed a dual-input deep learning framework, named AttnTAP, to improve the TCR-peptide binding prediction. It used the bi-directional long short-term memory model for robust feature extraction of TCR sequences, which considered the interrelationships between amino acids and their precursors and postcursors. We also introduced the attention mechanism to give amino acids different weights and pay more attention to the contributing ones. In addition, we used the multilayer perceptron model instead of complex networks to extract peptide features to reduce overfitting and computational costs. AttnTAP achieved high areas under the curves (AUCs) in TCR-peptide binding prediction on both balanced and unbalanced datasets (higher than 0.838 on McPAS-TCR and 0.908 on VDJdb). Furthermore, it had the highest average AUCs in TPP-I and TPP-II tasks compared with the other five popular models (TPP-I: 0.84 on McPAS-TCR and 0.894 on VDJdb; TPP-II: 0.837 on McPAS-TCR and 0.893 on VDJdb). In conclusion, AttnTAP is a reasonable and practical framework for predicting TCR-peptide binding, which can accelerate identifying neoantigens and activated T cells for immunotherapy to meet urgent clinical needs.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3389/fgene.2022.942491
- OA Status
- gold
- Cited By
- 19
- References
- 35
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4292566556
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4292566556Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3389/fgene.2022.942491Digital Object Identifier
- Title
-
AttnTAP: A Dual-input Framework Incorporating the Attention Mechanism for Accurately Predicting TCR-peptide BindingWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-08-22Full publication date if available
- Authors
-
Ying Xu, Xinyang Qian, Yao Tong, Fan Li, Ke Wang, Xuanping Zhang, Tao Liu, Jiayin WangList of authors in order
- Landing page
-
https://doi.org/10.3389/fgene.2022.942491Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.3389/fgene.2022.942491Direct OA link when available
- Concepts
-
Overfitting, T-cell receptor, Computational biology, Artificial intelligence, Computer science, Perceptron, Peptide, Artificial neural network, Amino acid, Machine learning, Biology, T cell, Biochemistry, Genetics, Immune systemTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
19Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 8, 2024: 8, 2023: 3Per-year citation counts (last 5 years)
- References (count)
-
35Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4292566556 |
|---|---|
| doi | https://doi.org/10.3389/fgene.2022.942491 |
| ids.doi | https://doi.org/10.3389/fgene.2022.942491 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/36072653 |
| ids.openalex | https://openalex.org/W4292566556 |
| fwci | 2.338314 |
| type | article |
| title | AttnTAP: A Dual-input Framework Incorporating the Attention Mechanism for Accurately Predicting TCR-peptide Binding |
| biblio.issue | |
| biblio.volume | 13 |
| biblio.last_page | 942491 |
| biblio.first_page | 942491 |
| topics[0].id | https://openalex.org/T12576 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | vaccines and immunoinformatics approaches |
| topics[1].id | https://openalex.org/T11103 |
| topics[1].field.id | https://openalex.org/fields/24 |
| topics[1].field.display_name | Immunology and Microbiology |
| topics[1].score | 0.9944999814033508 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2404 |
| topics[1].subfield.display_name | Microbiology |
| topics[1].display_name | Antimicrobial Peptides and Activities |
| topics[2].id | https://openalex.org/T10580 |
| topics[2].field.id | https://openalex.org/fields/24 |
| topics[2].field.display_name | Immunology and Microbiology |
| topics[2].score | 0.9936000108718872 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2403 |
| topics[2].subfield.display_name | Immunology |
| topics[2].display_name | Immunotherapy and Immune Responses |
| funders[0].id | https://openalex.org/F4320336567 |
| funders[0].ror | |
| funders[0].display_name | Natural Science Basic Research Program of Shaanxi Province |
| is_xpac | False |
| apc_list.value | 2950 |
| apc_list.currency | USD |
| apc_list.value_usd | 2950 |
| apc_paid.value | 2950 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2950 |
| concepts[0].id | https://openalex.org/C22019652 |
| concepts[0].level | 3 |
| concepts[0].score | 0.9030183553695679 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q331309 |
| concepts[0].display_name | Overfitting |
| concepts[1].id | https://openalex.org/C19317047 |
| concepts[1].level | 4 |
| concepts[1].score | 0.6957821249961853 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q412037 |
| concepts[1].display_name | T-cell receptor |
| concepts[2].id | https://openalex.org/C70721500 |
| concepts[2].level | 1 |
| concepts[2].score | 0.5750495195388794 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[2].display_name | Computational biology |
| concepts[3].id | https://openalex.org/C154945302 |
| concepts[3].level | 1 |
| concepts[3].score | 0.4947309195995331 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[3].display_name | Artificial intelligence |
| concepts[4].id | https://openalex.org/C41008148 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4884135127067566 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[4].display_name | Computer science |
| concepts[5].id | https://openalex.org/C60908668 |
| concepts[5].level | 3 |
| concepts[5].score | 0.48066937923431396 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q690207 |
| concepts[5].display_name | Perceptron |
| concepts[6].id | https://openalex.org/C2779281246 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4741482138633728 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q172847 |
| concepts[6].display_name | Peptide |
| concepts[7].id | https://openalex.org/C50644808 |
| concepts[7].level | 2 |
| concepts[7].score | 0.45077890157699585 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q192776 |
| concepts[7].display_name | Artificial neural network |
| concepts[8].id | https://openalex.org/C515207424 |
| concepts[8].level | 2 |
| concepts[8].score | 0.42965298891067505 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q8066 |
| concepts[8].display_name | Amino acid |
| concepts[9].id | https://openalex.org/C119857082 |
| concepts[9].level | 1 |
| concepts[9].score | 0.4250168800354004 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[9].display_name | Machine learning |
| concepts[10].id | https://openalex.org/C86803240 |
| concepts[10].level | 0 |
| concepts[10].score | 0.39671871066093445 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[10].display_name | Biology |
| concepts[11].id | https://openalex.org/C2776090121 |
| concepts[11].level | 3 |
| concepts[11].score | 0.22458496689796448 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q193529 |
| concepts[11].display_name | T cell |
| concepts[12].id | https://openalex.org/C55493867 |
| concepts[12].level | 1 |
| concepts[12].score | 0.1834317147731781 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[12].display_name | Biochemistry |
| concepts[13].id | https://openalex.org/C54355233 |
| concepts[13].level | 1 |
| concepts[13].score | 0.16834723949432373 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[13].display_name | Genetics |
| concepts[14].id | https://openalex.org/C8891405 |
| concepts[14].level | 2 |
| concepts[14].score | 0.16353130340576172 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q1059 |
| concepts[14].display_name | Immune system |
| keywords[0].id | https://openalex.org/keywords/overfitting |
| keywords[0].score | 0.9030183553695679 |
| keywords[0].display_name | Overfitting |
| keywords[1].id | https://openalex.org/keywords/t-cell-receptor |
| keywords[1].score | 0.6957821249961853 |
| keywords[1].display_name | T-cell receptor |
| keywords[2].id | https://openalex.org/keywords/computational-biology |
| keywords[2].score | 0.5750495195388794 |
| keywords[2].display_name | Computational biology |
| keywords[3].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[3].score | 0.4947309195995331 |
| keywords[3].display_name | Artificial intelligence |
| keywords[4].id | https://openalex.org/keywords/computer-science |
| keywords[4].score | 0.4884135127067566 |
| keywords[4].display_name | Computer science |
| keywords[5].id | https://openalex.org/keywords/perceptron |
| keywords[5].score | 0.48066937923431396 |
| keywords[5].display_name | Perceptron |
| keywords[6].id | https://openalex.org/keywords/peptide |
| keywords[6].score | 0.4741482138633728 |
| keywords[6].display_name | Peptide |
| keywords[7].id | https://openalex.org/keywords/artificial-neural-network |
| keywords[7].score | 0.45077890157699585 |
| keywords[7].display_name | Artificial neural network |
| keywords[8].id | https://openalex.org/keywords/amino-acid |
| keywords[8].score | 0.42965298891067505 |
| keywords[8].display_name | Amino acid |
| keywords[9].id | https://openalex.org/keywords/machine-learning |
| keywords[9].score | 0.4250168800354004 |
| keywords[9].display_name | Machine learning |
| keywords[10].id | https://openalex.org/keywords/biology |
| keywords[10].score | 0.39671871066093445 |
| keywords[10].display_name | Biology |
| keywords[11].id | https://openalex.org/keywords/t-cell |
| keywords[11].score | 0.22458496689796448 |
| keywords[11].display_name | T cell |
| keywords[12].id | https://openalex.org/keywords/biochemistry |
| keywords[12].score | 0.1834317147731781 |
| keywords[12].display_name | Biochemistry |
| keywords[13].id | https://openalex.org/keywords/genetics |
| keywords[13].score | 0.16834723949432373 |
| keywords[13].display_name | Genetics |
| keywords[14].id | https://openalex.org/keywords/immune-system |
| keywords[14].score | 0.16353130340576172 |
| keywords[14].display_name | Immune system |
| language | en |
| locations[0].id | doi:10.3389/fgene.2022.942491 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2597340651 |
| locations[0].source.issn | 1664-8021 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1664-8021 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Frontiers in Genetics |
| locations[0].source.host_organization | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_name | Frontiers Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_lineage_names | Frontiers Media |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Frontiers in Genetics |
| locations[0].landing_page_url | https://doi.org/10.3389/fgene.2022.942491 |
| locations[1].id | pmid:36072653 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Frontiers in genetics |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/36072653 |
| locations[2].id | pmh:oai:doaj.org/article:75eb91435f7641ebbd3adc8bca17789b |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Frontiers in Genetics, Vol 13 (2022) |
| locations[2].landing_page_url | https://doaj.org/article/75eb91435f7641ebbd3adc8bca17789b |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:9441555 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Front Genet |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/9441555 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5102815586 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-4211-4156 |
| authorships[0].author.display_name | Ying Xu |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I87445476 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[0].institutions[0].id | https://openalex.org/I87445476 |
| authorships[0].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Ying Xu |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[1].author.id | https://openalex.org/A5085869025 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Xinyang Qian |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I87445476 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[1].institutions[0].id | https://openalex.org/I87445476 |
| authorships[1].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Xinyang Qian |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[2].author.id | https://openalex.org/A5038569817 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-7885-3922 |
| authorships[2].author.display_name | Yao Tong |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I87445476 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[2].institutions[0].id | https://openalex.org/I87445476 |
| authorships[2].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Yao Tong |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[3].author.id | https://openalex.org/A5100373576 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-7566-1634 |
| authorships[3].author.display_name | Fan Li |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I87445476 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[3].institutions[0].id | https://openalex.org/I87445476 |
| authorships[3].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Fan Li |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[4].author.id | https://openalex.org/A5100360160 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-4508-1302 |
| authorships[4].author.display_name | Ke Wang |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].raw_affiliation_string | Geneplus Beijing Institute, Beijing, China |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I87445476 |
| authorships[4].affiliations[1].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[4].institutions[0].id | https://openalex.org/I87445476 |
| authorships[4].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Ke Wang |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China, Geneplus Beijing Institute, Beijing, China |
| authorships[5].author.id | https://openalex.org/A5115594780 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Xuanping Zhang |
| authorships[5].countries | CN |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I87445476 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[5].institutions[0].id | https://openalex.org/I87445476 |
| authorships[5].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Xuanping Zhang |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[6].author.id | https://openalex.org/A5100326853 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-6588-1051 |
| authorships[6].author.display_name | Tao Liu |
| authorships[6].countries | CN |
| authorships[6].affiliations[0].raw_affiliation_string | Geneplus Beijing Institute, Beijing, China |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I87445476 |
| authorships[6].affiliations[1].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[6].institutions[0].id | https://openalex.org/I87445476 |
| authorships[6].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[6].institutions[0].country_code | CN |
| authorships[6].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Tao Liu |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China, Geneplus Beijing Institute, Beijing, China |
| authorships[7].author.id | https://openalex.org/A5027360778 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-3862-6557 |
| authorships[7].author.display_name | Jiayin Wang |
| authorships[7].countries | CN |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I87445476 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| authorships[7].institutions[0].id | https://openalex.org/I87445476 |
| authorships[7].institutions[0].ror | https://ror.org/017zhmm22 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I87445476 |
| authorships[7].institutions[0].country_code | CN |
| authorships[7].institutions[0].display_name | Xi'an Jiaotong University |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Jiayin Wang |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | Department of Computer Science and Technology, School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an, China |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.3389/fgene.2022.942491 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | AttnTAP: A Dual-input Framework Incorporating the Attention Mechanism for Accurately Predicting TCR-peptide Binding |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12576 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | vaccines and immunoinformatics approaches |
| related_works | https://openalex.org/W1574414179, https://openalex.org/W4362597605, https://openalex.org/W3009056573, https://openalex.org/W2922073769, https://openalex.org/W4297676672, https://openalex.org/W4281702477, https://openalex.org/W4378510483, https://openalex.org/W4376166922, https://openalex.org/W2490526372, https://openalex.org/W2150029999 |
| cited_by_count | 19 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 8 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 8 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 3 |
| locations_count | 4 |
| best_oa_location.id | doi:10.3389/fgene.2022.942491 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2597340651 |
| best_oa_location.source.issn | 1664-8021 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1664-8021 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Frontiers in Genetics |
| best_oa_location.source.host_organization | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_name | Frontiers Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_lineage_names | Frontiers Media |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Frontiers in Genetics |
| best_oa_location.landing_page_url | https://doi.org/10.3389/fgene.2022.942491 |
| primary_location.id | doi:10.3389/fgene.2022.942491 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2597340651 |
| primary_location.source.issn | 1664-8021 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1664-8021 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Frontiers in Genetics |
| primary_location.source.host_organization | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_name | Frontiers Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_lineage_names | Frontiers Media |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Frontiers in Genetics |
| primary_location.landing_page_url | https://doi.org/10.3389/fgene.2022.942491 |
| publication_date | 2022-08-22 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W1501531009, https://openalex.org/W2979330275, https://openalex.org/W4297734170, https://openalex.org/W1551566597, https://openalex.org/W3082739140, https://openalex.org/W2158266834, https://openalex.org/W2670890989, https://openalex.org/W6681513673, https://openalex.org/W2143210482, https://openalex.org/W3040528450, https://openalex.org/W2797967938, https://openalex.org/W2919115771, https://openalex.org/W2901096054, https://openalex.org/W1614298861, https://openalex.org/W3196626346, https://openalex.org/W3113525558, https://openalex.org/W2043071551, https://openalex.org/W2146074801, https://openalex.org/W2041877620, https://openalex.org/W3080758869, https://openalex.org/W6674330103, https://openalex.org/W3116512470, https://openalex.org/W2613978126, https://openalex.org/W4200382517, https://openalex.org/W4385245566, https://openalex.org/W2737196618, https://openalex.org/W2165567657, https://openalex.org/W1964372737, https://openalex.org/W3196225703, https://openalex.org/W4223441615, https://openalex.org/W2939354476, https://openalex.org/W3012203069, https://openalex.org/W2517194566, https://openalex.org/W2095705004, https://openalex.org/W2143612262 |
| referenced_works_count | 35 |
| abstract_inverted_index.T | 312 |
| abstract_inverted_index.a | 124, 148, 296 |
| abstract_inverted_index.In | 55, 143, 208, 292 |
| abstract_inverted_index.It | 161 |
| abstract_inverted_index.We | 188 |
| abstract_inverted_index.be | 77 |
| abstract_inverted_index.by | 5, 80, 107 |
| abstract_inverted_index.in | 85, 94, 127, 238, 264 |
| abstract_inverted_index.is | 45, 96, 295 |
| abstract_inverted_index.it | 258 |
| abstract_inverted_index.of | 8, 13, 18, 26, 73, 138, 173, 217 |
| abstract_inverted_index.on | 242, 251, 255, 278, 282, 286, 290 |
| abstract_inverted_index.to | 22, 118, 134, 155, 194, 204, 220, 224, 316 |
| abstract_inverted_index.we | 146, 210 |
| abstract_inverted_index.Due | 21 |
| abstract_inverted_index.TCR | 53, 99, 174 |
| abstract_inverted_index.The | 89 |
| abstract_inverted_index.and | 30, 41, 140, 183, 186, 200, 227, 245, 253, 266, 280, 288, 298, 310 |
| abstract_inverted_index.are | 3 |
| abstract_inverted_index.can | 76, 306 |
| abstract_inverted_index.did | 113 |
| abstract_inverted_index.for | 38, 65, 98, 169, 301, 314 |
| abstract_inverted_index.had | 259 |
| abstract_inverted_index.not | 104, 114 |
| abstract_inverted_index.pay | 115, 201 |
| abstract_inverted_index.the | 16, 23, 35, 46, 70, 108, 119, 136, 157, 163, 178, 191, 205, 212, 235, 260, 271 |
| abstract_inverted_index.was | 103 |
| abstract_inverted_index.0.84 | 277 |
| abstract_inverted_index.AUCs | 263 |
| abstract_inverted_index.TCRs | 40, 95 |
| abstract_inverted_index.They | 111 |
| abstract_inverted_index.also | 112, 189 |
| abstract_inverted_index.been | 63 |
| abstract_inverted_index.both | 243 |
| abstract_inverted_index.deep | 150 |
| abstract_inverted_index.five | 273 |
| abstract_inverted_index.from | 49 |
| abstract_inverted_index.give | 195 |
| abstract_inverted_index.have | 62 |
| abstract_inverted_index.high | 232 |
| abstract_inverted_index.long | 165 |
| abstract_inverted_index.meet | 317 |
| abstract_inverted_index.more | 116, 202 |
| abstract_inverted_index.play | 123 |
| abstract_inverted_index.risk | 137 |
| abstract_inverted_index.role | 126 |
| abstract_inverted_index.some | 12 |
| abstract_inverted_index.than | 249 |
| abstract_inverted_index.that | 122 |
| abstract_inverted_index.this | 144 |
| abstract_inverted_index.used | 162, 211 |
| abstract_inverted_index.with | 270 |
| abstract_inverted_index.0.837 | 285 |
| abstract_inverted_index.0.838 | 250 |
| abstract_inverted_index.0.893 | 289 |
| abstract_inverted_index.0.894 | 281 |
| abstract_inverted_index.0.908 | 254 |
| abstract_inverted_index.TPP-I | 265 |
| abstract_inverted_index.acids | 93, 121, 182, 197 |
| abstract_inverted_index.amino | 92, 120, 181, 196 |
| abstract_inverted_index.areas | 233 |
| abstract_inverted_index.cells | 313 |
| abstract_inverted_index.flaws | 84 |
| abstract_inverted_index.model | 168, 215 |
| abstract_inverted_index.named | 153 |
| abstract_inverted_index.ones. | 207 |
| abstract_inverted_index.other | 272 |
| abstract_inverted_index.tasks | 268 |
| abstract_inverted_index.their | 42, 184 |
| abstract_inverted_index.these | 74 |
| abstract_inverted_index.under | 234 |
| abstract_inverted_index.which | 14, 102, 176, 305 |
| abstract_inverted_index.(AUCs) | 237 |
| abstract_inverted_index.(TCRs) | 2 |
| abstract_inverted_index.T-cell | 0, 27 |
| abstract_inverted_index.TPP-II | 267 |
| abstract_inverted_index.VDJdb; | 283 |
| abstract_inverted_index.and/or | 51 |
| abstract_inverted_index.costs. | 142, 229 |
| abstract_inverted_index.curves | 236 |
| abstract_inverted_index.formed | 4 |
| abstract_inverted_index.immune | 28 |
| abstract_inverted_index.memory | 167 |
| abstract_inverted_index.models | 275 |
| abstract_inverted_index.needs. | 320 |
| abstract_inverted_index.neural | 86 |
| abstract_inverted_index.random | 6 |
| abstract_inverted_index.recent | 56 |
| abstract_inverted_index.reduce | 225 |
| abstract_inverted_index.robust | 170 |
| abstract_inverted_index.study, | 145 |
| abstract_inverted_index.tended | 133 |
| abstract_inverted_index.urgent | 318 |
| abstract_inverted_index.years, | 57 |
| abstract_inverted_index.(TPP-I: | 276 |
| abstract_inverted_index.(higher | 248 |
| abstract_inverted_index.AttnTAP | 230, 294 |
| abstract_inverted_index.TPP-II: | 284 |
| abstract_inverted_index.VDJdb). | 256, 291 |
| abstract_inverted_index.antigen | 100 |
| abstract_inverted_index.average | 262 |
| abstract_inverted_index.between | 91, 180 |
| abstract_inverted_index.binding | 67, 159, 240 |
| abstract_inverted_index.complex | 131, 218 |
| abstract_inverted_index.design. | 88 |
| abstract_inverted_index.extract | 221 |
| abstract_inverted_index.feature | 171 |
| abstract_inverted_index.further | 78 |
| abstract_inverted_index.genomic | 9 |
| abstract_inverted_index.highest | 261 |
| abstract_inverted_index.improve | 156 |
| abstract_inverted_index.instead | 216 |
| abstract_inverted_index.limited | 31 |
| abstract_inverted_index.mediate | 15 |
| abstract_inverted_index.methods | 75 |
| abstract_inverted_index.network | 87 |
| abstract_inverted_index.peptide | 222 |
| abstract_inverted_index.popular | 274 |
| abstract_inverted_index.process | 25 |
| abstract_inverted_index.several | 58, 82 |
| abstract_inverted_index.weights | 199 |
| abstract_inverted_index.AttnTAP, | 154 |
| abstract_inverted_index.However, | 69 |
| abstract_inverted_index.achieved | 231 |
| abstract_inverted_index.balanced | 244 |
| abstract_inverted_index.binding, | 304 |
| abstract_inverted_index.clinical | 319 |
| abstract_inverted_index.compared | 269 |
| abstract_inverted_index.critical | 97 |
| abstract_inverted_index.datasets | 247 |
| abstract_inverted_index.existing | 109 |
| abstract_inverted_index.features | 223 |
| abstract_inverted_index.improved | 79 |
| abstract_inverted_index.increase | 135 |
| abstract_inverted_index.learning | 151 |
| abstract_inverted_index.methods. | 110 |
| abstract_inverted_index.networks | 132, 219 |
| abstract_inverted_index.peptides | 44 |
| abstract_inverted_index.properly | 105 |
| abstract_inverted_index.proposed | 64 |
| abstract_inverted_index.response | 29 |
| abstract_inverted_index.strategy | 37 |
| abstract_inverted_index.McPAS-TCR | 252, 279, 287 |
| abstract_inverted_index.Moreover, | 130 |
| abstract_inverted_index.activated | 311 |
| abstract_inverted_index.addition, | 209 |
| abstract_inverted_index.antigens. | 20 |
| abstract_inverted_index.attention | 117, 192, 203 |
| abstract_inverted_index.developed | 147 |
| abstract_inverted_index.different | 198 |
| abstract_inverted_index.elements, | 11 |
| abstract_inverted_index.empirical | 33 |
| abstract_inverted_index.evidence, | 34 |
| abstract_inverted_index.framework | 300 |
| abstract_inverted_index.mechanism | 193 |
| abstract_inverted_index.practical | 36, 299 |
| abstract_inverted_index.precursor | 10 |
| abstract_inverted_index.receptors | 1 |
| abstract_inverted_index.accelerate | 307 |
| abstract_inverted_index.approaches | 61 |
| abstract_inverted_index.biological | 32 |
| abstract_inverted_index.considered | 106, 177 |
| abstract_inverted_index.dual-input | 149 |
| abstract_inverted_index.extraction | 172 |
| abstract_inverted_index.framework, | 152 |
| abstract_inverted_index.individual | 52 |
| abstract_inverted_index.introduced | 190 |
| abstract_inverted_index.multilayer | 213 |
| abstract_inverted_index.overcoming | 81 |
| abstract_inverted_index.perceptron | 214 |
| abstract_inverted_index.population | 50 |
| abstract_inverted_index.precursors | 185 |
| abstract_inverted_index.predicting | 302 |
| abstract_inverted_index.prediction | 48, 241 |
| abstract_inverted_index.predictive | 71 |
| abstract_inverted_index.reasonable | 297 |
| abstract_inverted_index.recognized | 43 |
| abstract_inverted_index.sequences, | 175 |
| abstract_inverted_index.short-term | 166 |
| abstract_inverted_index.unbalanced | 246 |
| abstract_inverted_index.TCR-peptide | 66, 158, 239, 303 |
| abstract_inverted_index.complicated | 24 |
| abstract_inverted_index.conclusion, | 293 |
| abstract_inverted_index.identifying | 39, 308 |
| abstract_inverted_index.neoantigens | 309 |
| abstract_inverted_index.overfitting | 139, 226 |
| abstract_inverted_index.prediction. | 68, 160 |
| abstract_inverted_index.recognition | 17 |
| abstract_inverted_index.significant | 83, 125 |
| abstract_inverted_index.Furthermore, | 257 |
| abstract_inverted_index.contributing | 206 |
| abstract_inverted_index.machine/deep | 59 |
| abstract_inverted_index.performances | 72 |
| abstract_inverted_index.postcursors. | 187 |
| abstract_inverted_index.recognition, | 101 |
| abstract_inverted_index.repertoires. | 54 |
| abstract_inverted_index.specificity. | 129 |
| abstract_inverted_index.computational | 47, 141, 228 |
| abstract_inverted_index.immunotherapy | 315 |
| abstract_inverted_index.recombination | 7 |
| abstract_inverted_index.bi-directional | 164 |
| abstract_inverted_index.learning-based | 60 |
| abstract_inverted_index.antigen-binding | 128 |
| abstract_inverted_index.cancer-associated | 19 |
| abstract_inverted_index.interrelationship | 90 |
| abstract_inverted_index.interrelationships | 179 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 96 |
| corresponding_author_ids | https://openalex.org/A5027360778 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 8 |
| corresponding_institution_ids | https://openalex.org/I87445476 |
| citation_normalized_percentile.value | 0.85369313 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |