Bayesian mixture model for clustering rare-variant effects in human genetic studies Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.08.03.454967
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.1101/2021.08.03.454967
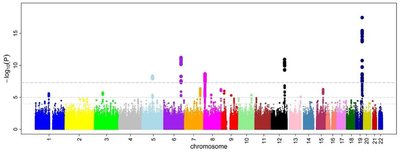
Rare-variant aggregate analysis from exome and whole genome sequencing data typically summarizes with a single statistic the signal for a gene or the unit that is being aggregated. However, when doing so, the effect profile within the unit may not be easily characterized across one or multiple phenotypes. Here, we present an approach we call Multiple Rarevariants and Phenotypes Mixture Model (MRPMM), which clusters rare variants into groups based on their effects on the multivariate phenotype and makes statistical inferences about the properties of the underlying mixture of genetic effects. Using summary statistic data from a meta-analysis of exome sequencing data of 184,698 individuals in the UK Biobank across 6 populations, we demonstrate that our mixture model can identify clusters of variants responsible for significantly disparate effects across a multivariate phenotype; we study three lipid and three renal traits separately. The method is able to estimate (1) the proportion of non-null variants, (2) whether variants with the same predicted consequence in one gene behave similarly, (3) whether variants across genes share effect profiles across the multivariate phenotype, and (4) whether different annotations differ in the magnitude of their effects. As rare-variant data and aggregation techniques become more common, this method can be used to ascribe further meaning to association results.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2021.08.03.454967
- https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdf
- OA Status
- green
- References
- 36
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3188604774
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3188604774Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2021.08.03.454967Digital Object Identifier
- Title
-
Bayesian mixture model for clustering rare-variant effects in human genetic studiesWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-08-05Full publication date if available
- Authors
-
Guhan Venkataraman, Yosuke Tanigawa, Matti Pirinen, Manuel A. RivasList of authors in order
- Landing page
-
https://doi.org/10.1101/2021.08.03.454967Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdfDirect OA link when available
- Concepts
-
Multivariate statistics, Biology, Computational biology, Biobank, Phenotype, Exome, Cluster analysis, Statistic, Genome-wide association study, Genetic association, Bayesian probability, Genetics, Exome sequencing, Gene, Statistics, Single-nucleotide polymorphism, Genotype, MathematicsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
36Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3188604774 |
|---|---|
| doi | https://doi.org/10.1101/2021.08.03.454967 |
| ids.doi | https://doi.org/10.1101/2021.08.03.454967 |
| ids.mag | 3188604774 |
| ids.openalex | https://openalex.org/W3188604774 |
| fwci | 0.0 |
| type | preprint |
| title | Bayesian mixture model for clustering rare-variant effects in human genetic studies |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10261 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic Associations and Epidemiology |
| topics[1].id | https://openalex.org/T11468 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9839000105857849 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genetic Mapping and Diversity in Plants and Animals |
| topics[2].id | https://openalex.org/T11764 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9828000068664551 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Evolution and Genetic Dynamics |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C161584116 |
| concepts[0].level | 2 |
| concepts[0].score | 0.6507934927940369 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1952580 |
| concepts[0].display_name | Multivariate statistics |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.5336098670959473 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C70721500 |
| concepts[2].level | 1 |
| concepts[2].score | 0.5248855948448181 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[2].display_name | Computational biology |
| concepts[3].id | https://openalex.org/C116567970 |
| concepts[3].level | 2 |
| concepts[3].score | 0.48771390318870544 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q864217 |
| concepts[3].display_name | Biobank |
| concepts[4].id | https://openalex.org/C127716648 |
| concepts[4].level | 3 |
| concepts[4].score | 0.4820740818977356 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q104053 |
| concepts[4].display_name | Phenotype |
| concepts[5].id | https://openalex.org/C10590036 |
| concepts[5].level | 5 |
| concepts[5].score | 0.461407333612442 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q417664 |
| concepts[5].display_name | Exome |
| concepts[6].id | https://openalex.org/C73555534 |
| concepts[6].level | 2 |
| concepts[6].score | 0.45037591457366943 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q622825 |
| concepts[6].display_name | Cluster analysis |
| concepts[7].id | https://openalex.org/C89128539 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4353913366794586 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1949963 |
| concepts[7].display_name | Statistic |
| concepts[8].id | https://openalex.org/C106208931 |
| concepts[8].level | 5 |
| concepts[8].score | 0.42970046401023865 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[8].display_name | Genome-wide association study |
| concepts[9].id | https://openalex.org/C186413461 |
| concepts[9].level | 5 |
| concepts[9].score | 0.423145592212677 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[9].display_name | Genetic association |
| concepts[10].id | https://openalex.org/C107673813 |
| concepts[10].level | 2 |
| concepts[10].score | 0.4146215319633484 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q812534 |
| concepts[10].display_name | Bayesian probability |
| concepts[11].id | https://openalex.org/C54355233 |
| concepts[11].level | 1 |
| concepts[11].score | 0.38476991653442383 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[11].display_name | Genetics |
| concepts[12].id | https://openalex.org/C16671776 |
| concepts[12].level | 4 |
| concepts[12].score | 0.3763321340084076 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q5420592 |
| concepts[12].display_name | Exome sequencing |
| concepts[13].id | https://openalex.org/C104317684 |
| concepts[13].level | 2 |
| concepts[13].score | 0.3666799068450928 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[13].display_name | Gene |
| concepts[14].id | https://openalex.org/C105795698 |
| concepts[14].level | 1 |
| concepts[14].score | 0.2657870054244995 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[14].display_name | Statistics |
| concepts[15].id | https://openalex.org/C153209595 |
| concepts[15].level | 4 |
| concepts[15].score | 0.258222758769989 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[15].display_name | Single-nucleotide polymorphism |
| concepts[16].id | https://openalex.org/C135763542 |
| concepts[16].level | 3 |
| concepts[16].score | 0.15463712811470032 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[16].display_name | Genotype |
| concepts[17].id | https://openalex.org/C33923547 |
| concepts[17].level | 0 |
| concepts[17].score | 0.1079200804233551 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[17].display_name | Mathematics |
| keywords[0].id | https://openalex.org/keywords/multivariate-statistics |
| keywords[0].score | 0.6507934927940369 |
| keywords[0].display_name | Multivariate statistics |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.5336098670959473 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/computational-biology |
| keywords[2].score | 0.5248855948448181 |
| keywords[2].display_name | Computational biology |
| keywords[3].id | https://openalex.org/keywords/biobank |
| keywords[3].score | 0.48771390318870544 |
| keywords[3].display_name | Biobank |
| keywords[4].id | https://openalex.org/keywords/phenotype |
| keywords[4].score | 0.4820740818977356 |
| keywords[4].display_name | Phenotype |
| keywords[5].id | https://openalex.org/keywords/exome |
| keywords[5].score | 0.461407333612442 |
| keywords[5].display_name | Exome |
| keywords[6].id | https://openalex.org/keywords/cluster-analysis |
| keywords[6].score | 0.45037591457366943 |
| keywords[6].display_name | Cluster analysis |
| keywords[7].id | https://openalex.org/keywords/statistic |
| keywords[7].score | 0.4353913366794586 |
| keywords[7].display_name | Statistic |
| keywords[8].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[8].score | 0.42970046401023865 |
| keywords[8].display_name | Genome-wide association study |
| keywords[9].id | https://openalex.org/keywords/genetic-association |
| keywords[9].score | 0.423145592212677 |
| keywords[9].display_name | Genetic association |
| keywords[10].id | https://openalex.org/keywords/bayesian-probability |
| keywords[10].score | 0.4146215319633484 |
| keywords[10].display_name | Bayesian probability |
| keywords[11].id | https://openalex.org/keywords/genetics |
| keywords[11].score | 0.38476991653442383 |
| keywords[11].display_name | Genetics |
| keywords[12].id | https://openalex.org/keywords/exome-sequencing |
| keywords[12].score | 0.3763321340084076 |
| keywords[12].display_name | Exome sequencing |
| keywords[13].id | https://openalex.org/keywords/gene |
| keywords[13].score | 0.3666799068450928 |
| keywords[13].display_name | Gene |
| keywords[14].id | https://openalex.org/keywords/statistics |
| keywords[14].score | 0.2657870054244995 |
| keywords[14].display_name | Statistics |
| keywords[15].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[15].score | 0.258222758769989 |
| keywords[15].display_name | Single-nucleotide polymorphism |
| keywords[16].id | https://openalex.org/keywords/genotype |
| keywords[16].score | 0.15463712811470032 |
| keywords[16].display_name | Genotype |
| keywords[17].id | https://openalex.org/keywords/mathematics |
| keywords[17].score | 0.1079200804233551 |
| keywords[17].display_name | Mathematics |
| language | en |
| locations[0].id | doi:10.1101/2021.08.03.454967 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2021.08.03.454967 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5053857674 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-0754-6631 |
| authorships[0].author.display_name | Guhan Venkataraman |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I97018004 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Biomedical Data Science, Stanford University, Stanford, CA, USA |
| authorships[0].institutions[0].id | https://openalex.org/I97018004 |
| authorships[0].institutions[0].ror | https://ror.org/00f54p054 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I97018004 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Stanford University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Guhan Ram Venkataraman |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Department of Biomedical Data Science, Stanford University, Stanford, CA, USA |
| authorships[1].author.id | https://openalex.org/A5003493606 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-9759-157X |
| authorships[1].author.display_name | Yosuke Tanigawa |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I97018004 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Biomedical Data Science, Stanford University, Stanford, CA, USA |
| authorships[1].institutions[0].id | https://openalex.org/I97018004 |
| authorships[1].institutions[0].ror | https://ror.org/00f54p054 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I97018004 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Stanford University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Yosuke Tanigawa |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Biomedical Data Science, Stanford University, Stanford, CA, USA |
| authorships[2].author.id | https://openalex.org/A5064582174 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-1664-1350 |
| authorships[2].author.display_name | Matti Pirinen |
| authorships[2].countries | FI |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I1323065761, https://openalex.org/I133731052 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Mathematics and Statistics, University of Helsinki, Helsinki, Finland |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I133731052, https://openalex.org/I4210107997, https://openalex.org/I4210133586, https://openalex.org/I4389424196 |
| authorships[2].affiliations[1].raw_affiliation_string | Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland |
| authorships[2].affiliations[2].institution_ids | https://openalex.org/I133731052 |
| authorships[2].affiliations[2].raw_affiliation_string | Department of Public Health, Faculty of Medicine, University of Helsinki, Helsinki, Finland |
| authorships[2].institutions[0].id | https://openalex.org/I4389424196 |
| authorships[2].institutions[0].ror | |
| authorships[2].institutions[0].type | |
| authorships[2].institutions[0].lineage | https://openalex.org/I4389424196 |
| authorships[2].institutions[0].country_code | |
| authorships[2].institutions[0].display_name | Deleted Institution |
| authorships[2].institutions[1].id | https://openalex.org/I4210107997 |
| authorships[2].institutions[1].ror | https://ror.org/014rks409 |
| authorships[2].institutions[1].type | education |
| authorships[2].institutions[1].lineage | https://openalex.org/I4210107997 |
| authorships[2].institutions[1].country_code | FI |
| authorships[2].institutions[1].display_name | Finland University |
| authorships[2].institutions[2].id | https://openalex.org/I4210133586 |
| authorships[2].institutions[2].ror | https://ror.org/030sbze61 |
| authorships[2].institutions[2].type | facility |
| authorships[2].institutions[2].lineage | https://openalex.org/I133731052, https://openalex.org/I4210133586 |
| authorships[2].institutions[2].country_code | FI |
| authorships[2].institutions[2].display_name | Institute for Molecular Medicine Finland |
| authorships[2].institutions[3].id | https://openalex.org/I1323065761 |
| authorships[2].institutions[3].ror | https://ror.org/01n1rg855 |
| authorships[2].institutions[3].type | other |
| authorships[2].institutions[3].lineage | https://openalex.org/I1323065761 |
| authorships[2].institutions[3].country_code | FI |
| authorships[2].institutions[3].display_name | Statistics Finland |
| authorships[2].institutions[4].id | https://openalex.org/I133731052 |
| authorships[2].institutions[4].ror | https://ror.org/040af2s02 |
| authorships[2].institutions[4].type | education |
| authorships[2].institutions[4].lineage | https://openalex.org/I133731052 |
| authorships[2].institutions[4].country_code | FI |
| authorships[2].institutions[4].display_name | University of Helsinki |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Matti Pirinen |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Mathematics and Statistics, University of Helsinki, Helsinki, Finland, Department of Public Health, Faculty of Medicine, University of Helsinki, Helsinki, Finland, Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland |
| authorships[3].author.id | https://openalex.org/A5073710029 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-1457-9925 |
| authorships[3].author.display_name | Manuel A. Rivas |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I97018004 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Biomedical Data Science, Stanford University, Stanford, CA, USA |
| authorships[3].institutions[0].id | https://openalex.org/I97018004 |
| authorships[3].institutions[0].ror | https://ror.org/00f54p054 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I97018004 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Stanford University |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Manuel A. Rivas |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Biomedical Data Science, Stanford University, Stanford, CA, USA |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Bayesian mixture model for clustering rare-variant effects in human genetic studies |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10261 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic Associations and Epidemiology |
| related_works | https://openalex.org/W2093932542, https://openalex.org/W2043256480, https://openalex.org/W2772505697, https://openalex.org/W3083501552, https://openalex.org/W2400191746, https://openalex.org/W2094815267, https://openalex.org/W1983406990, https://openalex.org/W2484239316, https://openalex.org/W3024198590, https://openalex.org/W4367844067 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2021.08.03.454967 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2021.08.03.454967 |
| primary_location.id | doi:10.1101/2021.08.03.454967 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2021/08/06/2021.08.03.454967.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2021.08.03.454967 |
| publication_date | 2021-08-05 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2020040634, https://openalex.org/W2171777347, https://openalex.org/W2097804771, https://openalex.org/W2096791516, https://openalex.org/W2163953557, https://openalex.org/W2063655230, https://openalex.org/W2007988561, https://openalex.org/W1971102564, https://openalex.org/W2070984858, https://openalex.org/W2100909778, https://openalex.org/W2159998163, https://openalex.org/W1850415666, https://openalex.org/W2168175751, https://openalex.org/W2110090977, https://openalex.org/W1843990841, https://openalex.org/W1990430353, https://openalex.org/W2302979210, https://openalex.org/W1579303078, https://openalex.org/W2020999234, https://openalex.org/W4231517135, https://openalex.org/W2138309709, https://openalex.org/W3121239749, https://openalex.org/W2104549677, https://openalex.org/W3011413344, https://openalex.org/W2522993044, https://openalex.org/W2034937344, https://openalex.org/W2128371599, https://openalex.org/W2133520037, https://openalex.org/W2233603983, https://openalex.org/W2949112115, https://openalex.org/W2885727518, https://openalex.org/W3185264067, https://openalex.org/W2163738067, https://openalex.org/W2417483443, https://openalex.org/W2129383830, https://openalex.org/W1991301405 |
| referenced_works_count | 36 |
| abstract_inverted_index.6 | 109 |
| abstract_inverted_index.a | 13, 19, 95, 128 |
| abstract_inverted_index.As | 189 |
| abstract_inverted_index.UK | 106 |
| abstract_inverted_index.an | 51 |
| abstract_inverted_index.be | 40, 201 |
| abstract_inverted_index.in | 104, 160, 183 |
| abstract_inverted_index.is | 25, 142 |
| abstract_inverted_index.of | 83, 87, 97, 101, 120, 149, 186 |
| abstract_inverted_index.on | 69, 72 |
| abstract_inverted_index.or | 21, 45 |
| abstract_inverted_index.to | 144, 203, 207 |
| abstract_inverted_index.we | 49, 53, 111, 131 |
| abstract_inverted_index.(1) | 146 |
| abstract_inverted_index.(2) | 152 |
| abstract_inverted_index.(3) | 165 |
| abstract_inverted_index.(4) | 178 |
| abstract_inverted_index.The | 140 |
| abstract_inverted_index.and | 5, 57, 76, 135, 177, 192 |
| abstract_inverted_index.can | 117, 200 |
| abstract_inverted_index.for | 18, 123 |
| abstract_inverted_index.may | 38 |
| abstract_inverted_index.not | 39 |
| abstract_inverted_index.one | 44, 161 |
| abstract_inverted_index.our | 114 |
| abstract_inverted_index.so, | 31 |
| abstract_inverted_index.the | 16, 22, 32, 36, 73, 81, 84, 105, 147, 156, 174, 184 |
| abstract_inverted_index.able | 143 |
| abstract_inverted_index.call | 54 |
| abstract_inverted_index.data | 9, 93, 100, 191 |
| abstract_inverted_index.from | 3, 94 |
| abstract_inverted_index.gene | 20, 162 |
| abstract_inverted_index.into | 66 |
| abstract_inverted_index.more | 196 |
| abstract_inverted_index.rare | 64 |
| abstract_inverted_index.same | 157 |
| abstract_inverted_index.that | 24, 113 |
| abstract_inverted_index.this | 198 |
| abstract_inverted_index.unit | 23, 37 |
| abstract_inverted_index.used | 202 |
| abstract_inverted_index.when | 29 |
| abstract_inverted_index.with | 12, 155 |
| abstract_inverted_index.Here, | 48 |
| abstract_inverted_index.Model | 60 |
| abstract_inverted_index.Using | 90 |
| abstract_inverted_index.about | 80 |
| abstract_inverted_index.based | 68 |
| abstract_inverted_index.being | 26 |
| abstract_inverted_index.doing | 30 |
| abstract_inverted_index.exome | 4, 98 |
| abstract_inverted_index.genes | 169 |
| abstract_inverted_index.lipid | 134 |
| abstract_inverted_index.makes | 77 |
| abstract_inverted_index.model | 116 |
| abstract_inverted_index.renal | 137 |
| abstract_inverted_index.share | 170 |
| abstract_inverted_index.study | 132 |
| abstract_inverted_index.their | 70, 187 |
| abstract_inverted_index.three | 133, 136 |
| abstract_inverted_index.which | 62 |
| abstract_inverted_index.whole | 6 |
| abstract_inverted_index.across | 43, 108, 127, 168, 173 |
| abstract_inverted_index.become | 195 |
| abstract_inverted_index.behave | 163 |
| abstract_inverted_index.differ | 182 |
| abstract_inverted_index.easily | 41 |
| abstract_inverted_index.effect | 33, 171 |
| abstract_inverted_index.genome | 7 |
| abstract_inverted_index.groups | 67 |
| abstract_inverted_index.method | 141, 199 |
| abstract_inverted_index.signal | 17 |
| abstract_inverted_index.single | 14 |
| abstract_inverted_index.traits | 138 |
| abstract_inverted_index.within | 35 |
| abstract_inverted_index.184,698 | 102 |
| abstract_inverted_index.Biobank | 107 |
| abstract_inverted_index.Mixture | 59 |
| abstract_inverted_index.ascribe | 204 |
| abstract_inverted_index.common, | 197 |
| abstract_inverted_index.effects | 71, 126 |
| abstract_inverted_index.further | 205 |
| abstract_inverted_index.genetic | 88 |
| abstract_inverted_index.meaning | 206 |
| abstract_inverted_index.mixture | 86, 115 |
| abstract_inverted_index.present | 50 |
| abstract_inverted_index.profile | 34 |
| abstract_inverted_index.summary | 91 |
| abstract_inverted_index.whether | 153, 166, 179 |
| abstract_inverted_index.(MRPMM), | 61 |
| abstract_inverted_index.However, | 28 |
| abstract_inverted_index.Multiple | 55 |
| abstract_inverted_index.analysis | 2 |
| abstract_inverted_index.approach | 52 |
| abstract_inverted_index.clusters | 63, 119 |
| abstract_inverted_index.effects. | 89, 188 |
| abstract_inverted_index.estimate | 145 |
| abstract_inverted_index.identify | 118 |
| abstract_inverted_index.multiple | 46 |
| abstract_inverted_index.non-null | 150 |
| abstract_inverted_index.profiles | 172 |
| abstract_inverted_index.results. | 209 |
| abstract_inverted_index.variants | 65, 121, 154, 167 |
| abstract_inverted_index.aggregate | 1 |
| abstract_inverted_index.different | 180 |
| abstract_inverted_index.disparate | 125 |
| abstract_inverted_index.magnitude | 185 |
| abstract_inverted_index.phenotype | 75 |
| abstract_inverted_index.predicted | 158 |
| abstract_inverted_index.statistic | 15, 92 |
| abstract_inverted_index.typically | 10 |
| abstract_inverted_index.variants, | 151 |
| abstract_inverted_index.Phenotypes | 58 |
| abstract_inverted_index.inferences | 79 |
| abstract_inverted_index.phenotype, | 176 |
| abstract_inverted_index.phenotype; | 130 |
| abstract_inverted_index.properties | 82 |
| abstract_inverted_index.proportion | 148 |
| abstract_inverted_index.sequencing | 8, 99 |
| abstract_inverted_index.similarly, | 164 |
| abstract_inverted_index.summarizes | 11 |
| abstract_inverted_index.techniques | 194 |
| abstract_inverted_index.underlying | 85 |
| abstract_inverted_index.aggregated. | 27 |
| abstract_inverted_index.aggregation | 193 |
| abstract_inverted_index.annotations | 181 |
| abstract_inverted_index.association | 208 |
| abstract_inverted_index.consequence | 159 |
| abstract_inverted_index.demonstrate | 112 |
| abstract_inverted_index.individuals | 103 |
| abstract_inverted_index.phenotypes. | 47 |
| abstract_inverted_index.responsible | 122 |
| abstract_inverted_index.separately. | 139 |
| abstract_inverted_index.statistical | 78 |
| abstract_inverted_index.Rare-variant | 0 |
| abstract_inverted_index.Rarevariants | 56 |
| abstract_inverted_index.multivariate | 74, 129, 175 |
| abstract_inverted_index.populations, | 110 |
| abstract_inverted_index.rare-variant | 190 |
| abstract_inverted_index.characterized | 42 |
| abstract_inverted_index.meta-analysis | 96 |
| abstract_inverted_index.significantly | 124 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5053857674 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 4 |
| corresponding_institution_ids | https://openalex.org/I97018004 |
| citation_normalized_percentile.value | 0.09964895 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |