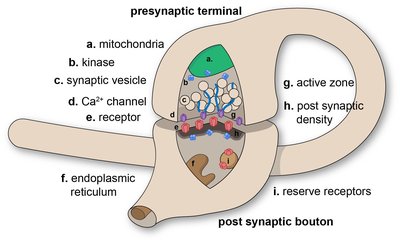
Ca2+ channel and active zone protein abundance intersects with input-specific synapse organization to shape functional synaptic diversity Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.7554/elife.88412
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.7554/elife.88412
Synaptic heterogeneity is a hallmark of nervous systems that enables complex and adaptable communication in neural circuits. To understand circuit function, it is thus critical to determine the factors that contribute to the functional diversity of synapses. We investigated the contributions of voltage-gated calcium channel (VGCC) abundance, spatial organization, and subunit composition to synapse diversity among and between synapses formed by two closely related Drosophila glutamatergic motor neurons with distinct neurotransmitter release probabilities (P r ). Surprisingly, VGCC levels are highly predictive of heterogeneous P r among individual synapses of either low- or high-P r inputs, but not between inputs. We find that the same number of VGCCs are more densely organized at high-P r synapses, consistent with tighter VGCC-synaptic vesicle coupling. We generated endogenously tagged lines to investigate VGCC subunits in vivo and found that the α2δ–3 subunit Straightjacket along with the CAST/ELKS active zone (AZ) protein Bruchpilot, both key regulators of VGCCs, are less abundant at high-P r inputs, yet positively correlate with P r among synapses formed by either input. Consistently, both Straightjacket and Bruchpilot levels are dynamically increased across AZs of both inputs when neurotransmitter release is potentiated to maintain stable communication following glutamate receptor inhibition. Together, these findings suggest a model in which VGCC and AZ protein abundance intersects with input-specific spatial and molecular organization to shape the functional diversity of synapses.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.7554/elife.88412
- OA Status
- gold
- Cited By
- 8
- References
- 109
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4384522776
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4384522776Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.7554/elife.88412Digital Object Identifier
- Title
-
Ca2+ channel and active zone protein abundance intersects with input-specific synapse organization to shape functional synaptic diversityWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-07-17Full publication date if available
- Authors
-
Audrey T. Medeiros, Scott J. Gratz, A. Delgado, Jason T. Ritt, Kate M. O’Connor-GilesList of authors in order
- Landing page
-
https://doi.org/10.7554/elife.88412Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.7554/elife.88412Direct OA link when available
- Concepts
-
Synapse, Abundance (ecology), Active zone, Neuroscience, Biology, Ecology, Synaptic vesicle, Genetics, Membrane, VesicleTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
8Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 6, 2024: 2Per-year citation counts (last 5 years)
- References (count)
-
109Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4384522776 |
|---|---|
| doi | https://doi.org/10.7554/elife.88412 |
| ids.doi | https://doi.org/10.7554/elife.88412 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39291956 |
| ids.openalex | https://openalex.org/W4384522776 |
| fwci | 1.68265562 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000818 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Animals |
| mesh[1].qualifier_ui | Q000378 |
| mesh[1].descriptor_ui | D013569 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | metabolism |
| mesh[1].descriptor_name | Synapses |
| mesh[2].qualifier_ui | Q000502 |
| mesh[2].descriptor_ui | D013569 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | physiology |
| mesh[2].descriptor_name | Synapses |
| mesh[3].qualifier_ui | Q000378 |
| mesh[3].descriptor_ui | D029721 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | metabolism |
| mesh[3].descriptor_name | Drosophila Proteins |
| mesh[4].qualifier_ui | Q000235 |
| mesh[4].descriptor_ui | D029721 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | genetics |
| mesh[4].descriptor_name | Drosophila Proteins |
| mesh[5].qualifier_ui | Q000378 |
| mesh[5].descriptor_ui | D015220 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | metabolism |
| mesh[5].descriptor_name | Calcium Channels |
| mesh[6].qualifier_ui | Q000378 |
| mesh[6].descriptor_ui | D009046 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | metabolism |
| mesh[6].descriptor_name | Motor Neurons |
| mesh[7].qualifier_ui | Q000502 |
| mesh[7].descriptor_ui | D009046 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | physiology |
| mesh[7].descriptor_name | Motor Neurons |
| mesh[8].qualifier_ui | Q000502 |
| mesh[8].descriptor_ui | D004330 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | physiology |
| mesh[8].descriptor_name | Drosophila |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D004331 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Drosophila melanogaster |
| mesh[10].qualifier_ui | Q000502 |
| mesh[10].descriptor_ui | D009435 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | physiology |
| mesh[10].descriptor_name | Synaptic Transmission |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000818 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Animals |
| mesh[12].qualifier_ui | Q000378 |
| mesh[12].descriptor_ui | D013569 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | metabolism |
| mesh[12].descriptor_name | Synapses |
| mesh[13].qualifier_ui | Q000502 |
| mesh[13].descriptor_ui | D013569 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | physiology |
| mesh[13].descriptor_name | Synapses |
| mesh[14].qualifier_ui | Q000378 |
| mesh[14].descriptor_ui | D029721 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | metabolism |
| mesh[14].descriptor_name | Drosophila Proteins |
| mesh[15].qualifier_ui | Q000235 |
| mesh[15].descriptor_ui | D029721 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | genetics |
| mesh[15].descriptor_name | Drosophila Proteins |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D015220 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Calcium Channels |
| mesh[17].qualifier_ui | Q000378 |
| mesh[17].descriptor_ui | D009046 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | metabolism |
| mesh[17].descriptor_name | Motor Neurons |
| mesh[18].qualifier_ui | Q000502 |
| mesh[18].descriptor_ui | D009046 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | physiology |
| mesh[18].descriptor_name | Motor Neurons |
| mesh[19].qualifier_ui | Q000502 |
| mesh[19].descriptor_ui | D004330 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | physiology |
| mesh[19].descriptor_name | Drosophila |
| mesh[20].qualifier_ui | Q000378 |
| mesh[20].descriptor_ui | D004331 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | metabolism |
| mesh[20].descriptor_name | Drosophila melanogaster |
| mesh[21].qualifier_ui | Q000502 |
| mesh[21].descriptor_ui | D009435 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | physiology |
| mesh[21].descriptor_name | Synaptic Transmission |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D000818 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Animals |
| mesh[23].qualifier_ui | Q000378 |
| mesh[23].descriptor_ui | D013569 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | metabolism |
| mesh[23].descriptor_name | Synapses |
| mesh[24].qualifier_ui | Q000502 |
| mesh[24].descriptor_ui | D013569 |
| mesh[24].is_major_topic | True |
| mesh[24].qualifier_name | physiology |
| mesh[24].descriptor_name | Synapses |
| mesh[25].qualifier_ui | Q000378 |
| mesh[25].descriptor_ui | D029721 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | metabolism |
| mesh[25].descriptor_name | Drosophila Proteins |
| mesh[26].qualifier_ui | Q000235 |
| mesh[26].descriptor_ui | D029721 |
| mesh[26].is_major_topic | True |
| mesh[26].qualifier_name | genetics |
| mesh[26].descriptor_name | Drosophila Proteins |
| mesh[27].qualifier_ui | Q000378 |
| mesh[27].descriptor_ui | D015220 |
| mesh[27].is_major_topic | True |
| mesh[27].qualifier_name | metabolism |
| mesh[27].descriptor_name | Calcium Channels |
| mesh[28].qualifier_ui | Q000378 |
| mesh[28].descriptor_ui | D009046 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | metabolism |
| mesh[28].descriptor_name | Motor Neurons |
| mesh[29].qualifier_ui | Q000502 |
| mesh[29].descriptor_ui | D009046 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | physiology |
| mesh[29].descriptor_name | Motor Neurons |
| mesh[30].qualifier_ui | Q000502 |
| mesh[30].descriptor_ui | D004330 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | physiology |
| mesh[30].descriptor_name | Drosophila |
| mesh[31].qualifier_ui | Q000378 |
| mesh[31].descriptor_ui | D004331 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | metabolism |
| mesh[31].descriptor_name | Drosophila melanogaster |
| mesh[32].qualifier_ui | Q000502 |
| mesh[32].descriptor_ui | D009435 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | physiology |
| mesh[32].descriptor_name | Synaptic Transmission |
| mesh[33].qualifier_ui | |
| mesh[33].descriptor_ui | D000818 |
| mesh[33].is_major_topic | False |
| mesh[33].qualifier_name | |
| mesh[33].descriptor_name | Animals |
| mesh[34].qualifier_ui | Q000378 |
| mesh[34].descriptor_ui | D013569 |
| mesh[34].is_major_topic | True |
| mesh[34].qualifier_name | metabolism |
| mesh[34].descriptor_name | Synapses |
| mesh[35].qualifier_ui | Q000502 |
| mesh[35].descriptor_ui | D013569 |
| mesh[35].is_major_topic | True |
| mesh[35].qualifier_name | physiology |
| mesh[35].descriptor_name | Synapses |
| mesh[36].qualifier_ui | Q000378 |
| mesh[36].descriptor_ui | D029721 |
| mesh[36].is_major_topic | True |
| mesh[36].qualifier_name | metabolism |
| mesh[36].descriptor_name | Drosophila Proteins |
| mesh[37].qualifier_ui | Q000235 |
| mesh[37].descriptor_ui | D029721 |
| mesh[37].is_major_topic | True |
| mesh[37].qualifier_name | genetics |
| mesh[37].descriptor_name | Drosophila Proteins |
| mesh[38].qualifier_ui | Q000378 |
| mesh[38].descriptor_ui | D015220 |
| mesh[38].is_major_topic | True |
| mesh[38].qualifier_name | metabolism |
| mesh[38].descriptor_name | Calcium Channels |
| mesh[39].qualifier_ui | Q000378 |
| mesh[39].descriptor_ui | D009046 |
| mesh[39].is_major_topic | False |
| mesh[39].qualifier_name | metabolism |
| mesh[39].descriptor_name | Motor Neurons |
| mesh[40].qualifier_ui | Q000502 |
| mesh[40].descriptor_ui | D009046 |
| mesh[40].is_major_topic | False |
| mesh[40].qualifier_name | physiology |
| mesh[40].descriptor_name | Motor Neurons |
| mesh[41].qualifier_ui | Q000502 |
| mesh[41].descriptor_ui | D004330 |
| mesh[41].is_major_topic | False |
| mesh[41].qualifier_name | physiology |
| mesh[41].descriptor_name | Drosophila |
| mesh[42].qualifier_ui | Q000378 |
| mesh[42].descriptor_ui | D004331 |
| mesh[42].is_major_topic | False |
| mesh[42].qualifier_name | metabolism |
| mesh[42].descriptor_name | Drosophila melanogaster |
| mesh[43].qualifier_ui | Q000502 |
| mesh[43].descriptor_ui | D009435 |
| mesh[43].is_major_topic | False |
| mesh[43].qualifier_name | physiology |
| mesh[43].descriptor_name | Synaptic Transmission |
| mesh[44].qualifier_ui | |
| mesh[44].descriptor_ui | D000818 |
| mesh[44].is_major_topic | False |
| mesh[44].qualifier_name | |
| mesh[44].descriptor_name | Animals |
| mesh[45].qualifier_ui | Q000378 |
| mesh[45].descriptor_ui | D013569 |
| mesh[45].is_major_topic | True |
| mesh[45].qualifier_name | metabolism |
| mesh[45].descriptor_name | Synapses |
| mesh[46].qualifier_ui | Q000502 |
| mesh[46].descriptor_ui | D013569 |
| mesh[46].is_major_topic | True |
| mesh[46].qualifier_name | physiology |
| mesh[46].descriptor_name | Synapses |
| mesh[47].qualifier_ui | Q000378 |
| mesh[47].descriptor_ui | D029721 |
| mesh[47].is_major_topic | True |
| mesh[47].qualifier_name | metabolism |
| mesh[47].descriptor_name | Drosophila Proteins |
| mesh[48].qualifier_ui | Q000235 |
| mesh[48].descriptor_ui | D029721 |
| mesh[48].is_major_topic | True |
| mesh[48].qualifier_name | genetics |
| mesh[48].descriptor_name | Drosophila Proteins |
| mesh[49].qualifier_ui | Q000378 |
| mesh[49].descriptor_ui | D015220 |
| mesh[49].is_major_topic | True |
| mesh[49].qualifier_name | metabolism |
| mesh[49].descriptor_name | Calcium Channels |
| type | article |
| title | Ca2+ channel and active zone protein abundance intersects with input-specific synapse organization to shape functional synaptic diversity |
| awards[0].id | https://openalex.org/G3111828781 |
| awards[0].funder_id | https://openalex.org/F4320337359 |
| awards[0].display_name | |
| awards[0].funder_award_id | F31NS122424 |
| awards[0].funder_display_name | National Institute of Neurological Disorders and Stroke |
| biblio.issue | |
| biblio.volume | 12 |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10077 |
| topics[0].field.id | https://openalex.org/fields/28 |
| topics[0].field.display_name | Neuroscience |
| topics[0].score | 0.998199999332428 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2804 |
| topics[0].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[0].display_name | Neuroscience and Neuropharmacology Research |
| topics[1].id | https://openalex.org/T10493 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9954000115394592 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Ion channel regulation and function |
| topics[2].id | https://openalex.org/T11601 |
| topics[2].field.id | https://openalex.org/fields/28 |
| topics[2].field.display_name | Neuroscience |
| topics[2].score | 0.9904000163078308 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2804 |
| topics[2].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[2].display_name | Neuroscience and Neural Engineering |
| funders[0].id | https://openalex.org/F4320337359 |
| funders[0].ror | https://ror.org/01s5ya894 |
| funders[0].display_name | National Institute of Neurological Disorders and Stroke |
| is_xpac | False |
| apc_list.value | 3000 |
| apc_list.currency | USD |
| apc_list.value_usd | 2500 |
| apc_paid.value | 3000 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2500 |
| concepts[0].id | https://openalex.org/C127445978 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7427308559417725 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q187181 |
| concepts[0].display_name | Synapse |
| concepts[1].id | https://openalex.org/C77077793 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5699795484542847 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q336019 |
| concepts[1].display_name | Abundance (ecology) |
| concepts[2].id | https://openalex.org/C187102610 |
| concepts[2].level | 5 |
| concepts[2].score | 0.5144813060760498 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q4677599 |
| concepts[2].display_name | Active zone |
| concepts[3].id | https://openalex.org/C169760540 |
| concepts[3].level | 1 |
| concepts[3].score | 0.4409835934638977 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q207011 |
| concepts[3].display_name | Neuroscience |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4262784421443939 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C18903297 |
| concepts[5].level | 1 |
| concepts[5].score | 0.20501145720481873 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7150 |
| concepts[5].display_name | Ecology |
| concepts[6].id | https://openalex.org/C148785051 |
| concepts[6].level | 4 |
| concepts[6].score | 0.1753970980644226 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q671136 |
| concepts[6].display_name | Synaptic vesicle |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.11045068502426147 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C41625074 |
| concepts[8].level | 2 |
| concepts[8].score | 0.0 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[8].display_name | Membrane |
| concepts[9].id | https://openalex.org/C130316041 |
| concepts[9].level | 3 |
| concepts[9].score | 0.0 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q189206 |
| concepts[9].display_name | Vesicle |
| keywords[0].id | https://openalex.org/keywords/synapse |
| keywords[0].score | 0.7427308559417725 |
| keywords[0].display_name | Synapse |
| keywords[1].id | https://openalex.org/keywords/abundance |
| keywords[1].score | 0.5699795484542847 |
| keywords[1].display_name | Abundance (ecology) |
| keywords[2].id | https://openalex.org/keywords/active-zone |
| keywords[2].score | 0.5144813060760498 |
| keywords[2].display_name | Active zone |
| keywords[3].id | https://openalex.org/keywords/neuroscience |
| keywords[3].score | 0.4409835934638977 |
| keywords[3].display_name | Neuroscience |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.4262784421443939 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/ecology |
| keywords[5].score | 0.20501145720481873 |
| keywords[5].display_name | Ecology |
| keywords[6].id | https://openalex.org/keywords/synaptic-vesicle |
| keywords[6].score | 0.1753970980644226 |
| keywords[6].display_name | Synaptic vesicle |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.11045068502426147 |
| keywords[7].display_name | Genetics |
| language | en |
| locations[0].id | doi:10.7554/elife.88412 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S1336409049 |
| locations[0].source.issn | 2050-084X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2050-084X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | eLife |
| locations[0].source.host_organization | https://openalex.org/P4310311710 |
| locations[0].source.host_organization_name | eLife Sciences Publications Ltd |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310311710 |
| locations[0].source.host_organization_lineage_names | eLife Sciences Publications Ltd |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | eLife |
| locations[0].landing_page_url | https://doi.org/10.7554/elife.88412 |
| locations[1].id | pmid:39291956 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | eLife |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39291956 |
| locations[2].id | pmh:oai:doaj.org/article:d58d6cd12aaa4570b7a5d9353593b7e3 |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | eLife, Vol 12 (2024) |
| locations[2].landing_page_url | https://doaj.org/article/d58d6cd12aaa4570b7a5d9353593b7e3 |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:11410372 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | eLife |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11410372 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5025415939 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-5562-4772 |
| authorships[0].author.display_name | Audrey T. Medeiros |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I175594653 |
| authorships[0].affiliations[0].raw_affiliation_string | Neuroscience Graduate Training Program, Brown University |
| authorships[0].institutions[0].id | https://openalex.org/I175594653 |
| authorships[0].institutions[0].ror | https://ror.org/02ct41q97 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I175594653 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | John Brown University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Audrey T Medeiros |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Neuroscience Graduate Training Program, Brown University |
| authorships[1].author.id | https://openalex.org/A5040831583 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-0106-8336 |
| authorships[1].author.display_name | Scott J. Gratz |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I175594653 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Neuroscience, Brown University |
| authorships[1].institutions[0].id | https://openalex.org/I175594653 |
| authorships[1].institutions[0].ror | https://ror.org/02ct41q97 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I175594653 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | John Brown University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Scott J Gratz |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Neuroscience, Brown University |
| authorships[2].author.id | https://openalex.org/A5090504612 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-9712-0305 |
| authorships[2].author.display_name | A. Delgado |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I175594653 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Neuroscience, Brown University |
| authorships[2].institutions[0].id | https://openalex.org/I175594653 |
| authorships[2].institutions[0].ror | https://ror.org/02ct41q97 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I175594653 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | John Brown University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Ambar Delgado |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Neuroscience, Brown University |
| authorships[3].author.id | https://openalex.org/A5090402475 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-3113-7977 |
| authorships[3].author.display_name | Jason T. Ritt |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I175594653 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Neuroscience, Brown University |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I1314596251, https://openalex.org/I27804330 |
| authorships[3].affiliations[1].raw_affiliation_string | Carney Institute for Brain Science, Brown University |
| authorships[3].institutions[0].id | https://openalex.org/I1314596251 |
| authorships[3].institutions[0].ror | https://ror.org/00dcv1019 |
| authorships[3].institutions[0].type | facility |
| authorships[3].institutions[0].lineage | https://openalex.org/I1314596251, https://openalex.org/I4210140341 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Allen Institute for Brain Science |
| authorships[3].institutions[1].id | https://openalex.org/I27804330 |
| authorships[3].institutions[1].ror | https://ror.org/05gq02987 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I27804330 |
| authorships[3].institutions[1].country_code | US |
| authorships[3].institutions[1].display_name | Brown University |
| authorships[3].institutions[2].id | https://openalex.org/I175594653 |
| authorships[3].institutions[2].ror | https://ror.org/02ct41q97 |
| authorships[3].institutions[2].type | education |
| authorships[3].institutions[2].lineage | https://openalex.org/I175594653 |
| authorships[3].institutions[2].country_code | US |
| authorships[3].institutions[2].display_name | John Brown University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jason T Ritt |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Carney Institute for Brain Science, Brown University, Department of Neuroscience, Brown University |
| authorships[4].author.id | https://openalex.org/A5082368830 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-2259-8408 |
| authorships[4].author.display_name | Kate M. O’Connor-Giles |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I175594653 |
| authorships[4].affiliations[0].raw_affiliation_string | Neuroscience Graduate Training Program, Brown University |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I175594653 |
| authorships[4].affiliations[1].raw_affiliation_string | Department of Neuroscience, Brown University |
| authorships[4].affiliations[2].institution_ids | https://openalex.org/I1314596251, https://openalex.org/I27804330 |
| authorships[4].affiliations[2].raw_affiliation_string | Carney Institute for Brain Science, Brown University |
| authorships[4].institutions[0].id | https://openalex.org/I1314596251 |
| authorships[4].institutions[0].ror | https://ror.org/00dcv1019 |
| authorships[4].institutions[0].type | facility |
| authorships[4].institutions[0].lineage | https://openalex.org/I1314596251, https://openalex.org/I4210140341 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Allen Institute for Brain Science |
| authorships[4].institutions[1].id | https://openalex.org/I27804330 |
| authorships[4].institutions[1].ror | https://ror.org/05gq02987 |
| authorships[4].institutions[1].type | education |
| authorships[4].institutions[1].lineage | https://openalex.org/I27804330 |
| authorships[4].institutions[1].country_code | US |
| authorships[4].institutions[1].display_name | Brown University |
| authorships[4].institutions[2].id | https://openalex.org/I175594653 |
| authorships[4].institutions[2].ror | https://ror.org/02ct41q97 |
| authorships[4].institutions[2].type | education |
| authorships[4].institutions[2].lineage | https://openalex.org/I175594653 |
| authorships[4].institutions[2].country_code | US |
| authorships[4].institutions[2].display_name | John Brown University |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Kate M O'Connor-Giles |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Carney Institute for Brain Science, Brown University, Department of Neuroscience, Brown University, Neuroscience Graduate Training Program, Brown University |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.7554/elife.88412 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2023-07-18T00:00:00 |
| display_name | Ca2+ channel and active zone protein abundance intersects with input-specific synapse organization to shape functional synaptic diversity |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10077 |
| primary_topic.field.id | https://openalex.org/fields/28 |
| primary_topic.field.display_name | Neuroscience |
| primary_topic.score | 0.998199999332428 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2804 |
| primary_topic.subfield.display_name | Cellular and Molecular Neuroscience |
| primary_topic.display_name | Neuroscience and Neuropharmacology Research |
| related_works | https://openalex.org/W4220770200, https://openalex.org/W2149270136, https://openalex.org/W2982288654, https://openalex.org/W2991375491, https://openalex.org/W2080611091, https://openalex.org/W2030092367, https://openalex.org/W2029292398, https://openalex.org/W4233339657, https://openalex.org/W3036204849, https://openalex.org/W753497295 |
| cited_by_count | 8 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 6 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 4 |
| best_oa_location.id | doi:10.7554/elife.88412 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S1336409049 |
| best_oa_location.source.issn | 2050-084X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2050-084X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | eLife |
| best_oa_location.source.host_organization | https://openalex.org/P4310311710 |
| best_oa_location.source.host_organization_name | eLife Sciences Publications Ltd |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310311710 |
| best_oa_location.source.host_organization_lineage_names | eLife Sciences Publications Ltd |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | eLife |
| best_oa_location.landing_page_url | https://doi.org/10.7554/elife.88412 |
| primary_location.id | doi:10.7554/elife.88412 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S1336409049 |
| primary_location.source.issn | 2050-084X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2050-084X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | eLife |
| primary_location.source.host_organization | https://openalex.org/P4310311710 |
| primary_location.source.host_organization_name | eLife Sciences Publications Ltd |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310311710 |
| primary_location.source.host_organization_lineage_names | eLife Sciences Publications Ltd |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | eLife |
| primary_location.landing_page_url | https://doi.org/10.7554/elife.88412 |
| publication_date | 2023-07-17 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2822028304, https://openalex.org/W4306748254, https://openalex.org/W3120962416, https://openalex.org/W3039428993, https://openalex.org/W2088605279, https://openalex.org/W2084840477, https://openalex.org/W1972529694, https://openalex.org/W2027905035, https://openalex.org/W2512669502, https://openalex.org/W2903612916, https://openalex.org/W2005686453, https://openalex.org/W2569441742, https://openalex.org/W1600089255, https://openalex.org/W2004077897, https://openalex.org/W2035488746, https://openalex.org/W2061619102, https://openalex.org/W4386386535, https://openalex.org/W4285494590, https://openalex.org/W1999369847, https://openalex.org/W4311356977, https://openalex.org/W2135286498, https://openalex.org/W2055467158, https://openalex.org/W2901386532, https://openalex.org/W3010654069, https://openalex.org/W2881950577, https://openalex.org/W2091694654, https://openalex.org/W2014786857, https://openalex.org/W2028789389, https://openalex.org/W2914408698, https://openalex.org/W2151889253, https://openalex.org/W2145791439, https://openalex.org/W2098341193, https://openalex.org/W3176946842, https://openalex.org/W2799399472, https://openalex.org/W2985462421, https://openalex.org/W4321185632, https://openalex.org/W2154395280, https://openalex.org/W2914883871, https://openalex.org/W2129788878, https://openalex.org/W1970729216, https://openalex.org/W2027014007, https://openalex.org/W1994057447, https://openalex.org/W2032313226, https://openalex.org/W4377711803, https://openalex.org/W3048652890, https://openalex.org/W2409498041, https://openalex.org/W2093514926, https://openalex.org/W2994539845, https://openalex.org/W2013077952, https://openalex.org/W1939841200, https://openalex.org/W2953287524, https://openalex.org/W4386071242, https://openalex.org/W2066439481, https://openalex.org/W4321317618, https://openalex.org/W2102548112, https://openalex.org/W2055518516, https://openalex.org/W1493682555, https://openalex.org/W2779285120, https://openalex.org/W2802878745, https://openalex.org/W2136326277, https://openalex.org/W2912866984, https://openalex.org/W2057761743, https://openalex.org/W2064575171, https://openalex.org/W2116601957, https://openalex.org/W4210283200, https://openalex.org/W2008157904, https://openalex.org/W2134178604, https://openalex.org/W2514973692, https://openalex.org/W2054214388, https://openalex.org/W2042108686, https://openalex.org/W3100679542, https://openalex.org/W4388694045, https://openalex.org/W2093812780, https://openalex.org/W2625392953, https://openalex.org/W3203020648, https://openalex.org/W2140378892, https://openalex.org/W2245299918, https://openalex.org/W1970899379, https://openalex.org/W2030428080, https://openalex.org/W2592481914, https://openalex.org/W4205262694, https://openalex.org/W4375948266, https://openalex.org/W4391424688, https://openalex.org/W1550492084, https://openalex.org/W1980384788, https://openalex.org/W2007239742, https://openalex.org/W2155758917, https://openalex.org/W2091649989, https://openalex.org/W2065178022, https://openalex.org/W3016750913, https://openalex.org/W2977087614, https://openalex.org/W2753189667, https://openalex.org/W2887311672, https://openalex.org/W2884522219, https://openalex.org/W3209711083, https://openalex.org/W3146403365, https://openalex.org/W1983681582, https://openalex.org/W2103424071, https://openalex.org/W2291883125, https://openalex.org/W2729112625, https://openalex.org/W2548069103, https://openalex.org/W2088724185, https://openalex.org/W2519514824, https://openalex.org/W2581013199, https://openalex.org/W2027829143, https://openalex.org/W2990990666, https://openalex.org/W4388459721, https://openalex.org/W4206971710, https://openalex.org/W2013895597 |
| referenced_works_count | 109 |
| abstract_inverted_index.P | 84, 165 |
| abstract_inverted_index.a | 3, 204 |
| abstract_inverted_index.r | 74, 85, 94, 114, 159, 166 |
| abstract_inverted_index.(P | 73 |
| abstract_inverted_index.). | 75 |
| abstract_inverted_index.AZ | 210 |
| abstract_inverted_index.To | 17 |
| abstract_inverted_index.We | 37, 100, 122 |
| abstract_inverted_index.at | 112, 157 |
| abstract_inverted_index.by | 60, 170 |
| abstract_inverted_index.in | 14, 131, 206 |
| abstract_inverted_index.is | 2, 22, 190 |
| abstract_inverted_index.it | 21 |
| abstract_inverted_index.of | 5, 35, 41, 82, 89, 106, 152, 184, 225 |
| abstract_inverted_index.or | 92 |
| abstract_inverted_index.to | 25, 31, 52, 127, 192, 220 |
| abstract_inverted_index.AZs | 183 |
| abstract_inverted_index.and | 11, 49, 56, 133, 176, 209, 217 |
| abstract_inverted_index.are | 79, 108, 154, 179 |
| abstract_inverted_index.but | 96 |
| abstract_inverted_index.key | 150 |
| abstract_inverted_index.not | 97 |
| abstract_inverted_index.the | 27, 32, 39, 103, 136, 142, 222 |
| abstract_inverted_index.two | 61 |
| abstract_inverted_index.yet | 161 |
| abstract_inverted_index.(AZ) | 146 |
| abstract_inverted_index.VGCC | 77, 129, 208 |
| abstract_inverted_index.both | 149, 174, 185 |
| abstract_inverted_index.find | 101 |
| abstract_inverted_index.less | 155 |
| abstract_inverted_index.low- | 91 |
| abstract_inverted_index.more | 109 |
| abstract_inverted_index.same | 104 |
| abstract_inverted_index.that | 8, 29, 102, 135 |
| abstract_inverted_index.thus | 23 |
| abstract_inverted_index.vivo | 132 |
| abstract_inverted_index.when | 187 |
| abstract_inverted_index.with | 68, 117, 141, 164, 214 |
| abstract_inverted_index.zone | 145 |
| abstract_inverted_index.VGCCs | 107 |
| abstract_inverted_index.along | 140 |
| abstract_inverted_index.among | 55, 86, 167 |
| abstract_inverted_index.found | 134 |
| abstract_inverted_index.lines | 126 |
| abstract_inverted_index.model | 205 |
| abstract_inverted_index.motor | 66 |
| abstract_inverted_index.shape | 221 |
| abstract_inverted_index.these | 201 |
| abstract_inverted_index.which | 207 |
| abstract_inverted_index.(VGCC) | 45 |
| abstract_inverted_index.VGCCs, | 153 |
| abstract_inverted_index.across | 182 |
| abstract_inverted_index.active | 144 |
| abstract_inverted_index.either | 90, 171 |
| abstract_inverted_index.formed | 59, 169 |
| abstract_inverted_index.high-P | 93, 113, 158 |
| abstract_inverted_index.highly | 80 |
| abstract_inverted_index.input. | 172 |
| abstract_inverted_index.inputs | 186 |
| abstract_inverted_index.levels | 78, 178 |
| abstract_inverted_index.neural | 15 |
| abstract_inverted_index.number | 105 |
| abstract_inverted_index.stable | 194 |
| abstract_inverted_index.tagged | 125 |
| abstract_inverted_index.between | 57, 98 |
| abstract_inverted_index.calcium | 43 |
| abstract_inverted_index.channel | 44 |
| abstract_inverted_index.circuit | 19 |
| abstract_inverted_index.closely | 62 |
| abstract_inverted_index.complex | 10 |
| abstract_inverted_index.densely | 110 |
| abstract_inverted_index.enables | 9 |
| abstract_inverted_index.factors | 28 |
| abstract_inverted_index.inputs, | 95, 160 |
| abstract_inverted_index.inputs. | 99 |
| abstract_inverted_index.nervous | 6 |
| abstract_inverted_index.neurons | 67 |
| abstract_inverted_index.protein | 147, 211 |
| abstract_inverted_index.related | 63 |
| abstract_inverted_index.release | 71, 189 |
| abstract_inverted_index.spatial | 47, 216 |
| abstract_inverted_index.subunit | 50, 138 |
| abstract_inverted_index.suggest | 203 |
| abstract_inverted_index.synapse | 53 |
| abstract_inverted_index.systems | 7 |
| abstract_inverted_index.tighter | 118 |
| abstract_inverted_index.vesicle | 120 |
| abstract_inverted_index.Synaptic | 0 |
| abstract_inverted_index.abundant | 156 |
| abstract_inverted_index.critical | 24 |
| abstract_inverted_index.distinct | 69 |
| abstract_inverted_index.findings | 202 |
| abstract_inverted_index.hallmark | 4 |
| abstract_inverted_index.maintain | 193 |
| abstract_inverted_index.receptor | 198 |
| abstract_inverted_index.subunits | 130 |
| abstract_inverted_index.synapses | 58, 88, 168 |
| abstract_inverted_index.CAST/ELKS | 143 |
| abstract_inverted_index.Together, | 200 |
| abstract_inverted_index.abundance | 212 |
| abstract_inverted_index.adaptable | 12 |
| abstract_inverted_index.circuits. | 16 |
| abstract_inverted_index.correlate | 163 |
| abstract_inverted_index.coupling. | 121 |
| abstract_inverted_index.determine | 26 |
| abstract_inverted_index.diversity | 34, 54, 224 |
| abstract_inverted_index.following | 196 |
| abstract_inverted_index.function, | 20 |
| abstract_inverted_index.generated | 123 |
| abstract_inverted_index.glutamate | 197 |
| abstract_inverted_index.increased | 181 |
| abstract_inverted_index.molecular | 218 |
| abstract_inverted_index.organized | 111 |
| abstract_inverted_index.synapses, | 115 |
| abstract_inverted_index.synapses. | 36, 226 |
| abstract_inverted_index.α2δ–3 | 137 |
| abstract_inverted_index.Bruchpilot | 177 |
| abstract_inverted_index.Drosophila | 64 |
| abstract_inverted_index.abundance, | 46 |
| abstract_inverted_index.consistent | 116 |
| abstract_inverted_index.contribute | 30 |
| abstract_inverted_index.functional | 33, 223 |
| abstract_inverted_index.individual | 87 |
| abstract_inverted_index.intersects | 213 |
| abstract_inverted_index.positively | 162 |
| abstract_inverted_index.predictive | 81 |
| abstract_inverted_index.regulators | 151 |
| abstract_inverted_index.understand | 18 |
| abstract_inverted_index.Bruchpilot, | 148 |
| abstract_inverted_index.composition | 51 |
| abstract_inverted_index.dynamically | 180 |
| abstract_inverted_index.inhibition. | 199 |
| abstract_inverted_index.investigate | 128 |
| abstract_inverted_index.potentiated | 191 |
| abstract_inverted_index.endogenously | 124 |
| abstract_inverted_index.investigated | 38 |
| abstract_inverted_index.organization | 219 |
| abstract_inverted_index.Consistently, | 173 |
| abstract_inverted_index.Surprisingly, | 76 |
| abstract_inverted_index.VGCC-synaptic | 119 |
| abstract_inverted_index.communication | 13, 195 |
| abstract_inverted_index.contributions | 40 |
| abstract_inverted_index.glutamatergic | 65 |
| abstract_inverted_index.heterogeneity | 1 |
| abstract_inverted_index.heterogeneous | 83 |
| abstract_inverted_index.organization, | 48 |
| abstract_inverted_index.probabilities | 72 |
| abstract_inverted_index.voltage-gated | 42 |
| abstract_inverted_index.Straightjacket | 139, 175 |
| abstract_inverted_index.input-specific | 215 |
| abstract_inverted_index.neurotransmitter | 70, 188 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 94 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| citation_normalized_percentile.value | 0.80759554 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |