Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias Article Swipe
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1093/hmg/ddy263
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1093/hmg/ddy263
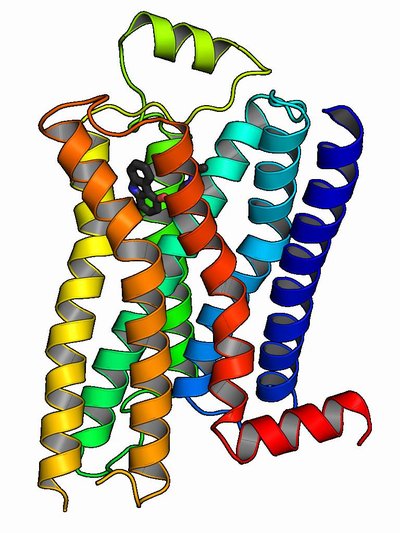
The calcium-sensing receptor (CaSR) is a homodimeric G-protein-coupled receptor that signals via intracellular calcium (Ca2+i) mobilisation and phosphorylation of extracellular signal-regulated kinase 1/2 (ERK) to regulate extracellular calcium (Ca2+e) homeostasis. The central importance of the CaSR in Ca2+e homeostasis has been demonstrated by the identification of loss- or gain-of-function CaSR mutations that lead to familial hypocalciuric hypercalcaemia (FHH) or autosomal dominant hypocalcaemia (ADH), respectively. However, the mechanisms determining whether the CaSR signals via Ca2+i or ERK have not been established, and we hypothesised that some CaSR residues, which are the site of both loss- and gain-of-function mutations, may act as molecular switches to direct signalling through these pathways. An analysis of CaSR mutations identified in >300 hypercalcaemic and hypocalcaemic probands revealed five 'disease-switch' residues (Gln27, Asn178, Ser657, Ser820 and Thr828) that are affected by FHH and ADH mutations. Functional expression studies using HEK293 cells showed disease-switch residue mutations to commonly display signalling bias. For example, two FHH-associated mutations (p.Asn178Asp and p.Ser820Ala) impaired Ca2+i signalling without altering ERK phosphorylation. In contrast, an ADH-associated p.Ser657Cys mutation uncoupled signalling by leading to increased Ca2+i mobilization while decreasing ERK phosphorylation. Structural analysis of these five CaSR disease-switch residues together with four reported disease-switch residues revealed these residues to be located at conformationally active regions of the CaSR such as the extracellular dimer interface and transmembrane domain. Thus, our findings indicate that disease-switch residues are located at sites critical for CaSR activation and play a role in mediating signalling bias.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1093/hmg/ddy263
- https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdf
- OA Status
- hybrid
- Cited By
- 28
- References
- 58
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2884306505
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2884306505Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1093/hmg/ddy263Digital Object Identifier
- Title
-
Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling biasWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2018Year of publication
- Publication date
-
2018-07-19Full publication date if available
- Authors
-
Caroline M. Gorvin, Morten Frost, Tomas Malinauskas, Treena Cranston, Hannah Boon, Christian Siebold, E. Yvonne Jones, Fadil Hannan, Rajesh V. ThakkerList of authors in order
- Landing page
-
https://doi.org/10.1093/hmg/ddy263Publisher landing page
- PDF URL
-
https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdfDirect OA link when available
- Concepts
-
Calcium-sensing receptor, Biology, Extracellular, Phosphorylation, Receptor, Mutation, MAPK/ERK pathway, Signal transduction, Cell biology, G protein-coupled receptor, Calcium, Calcium metabolism, Biochemistry, Internal medicine, Gene, MedicineTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
28Total citation count in OpenAlex
- Citations by year (recent)
-
2024: 5, 2023: 2, 2022: 2, 2021: 7, 2020: 7Per-year citation counts (last 5 years)
- References (count)
-
58Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2884306505 |
|---|---|
| doi | https://doi.org/10.1093/hmg/ddy263 |
| ids.doi | https://doi.org/10.1093/hmg/ddy263 |
| ids.mag | 2884306505 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/30052933 |
| ids.openalex | https://openalex.org/W2884306505 |
| fwci | 2.03498763 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000595 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Amino Acid Sequence |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D020013 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Calcium Signaling |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D004252 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | DNA Mutational Analysis |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D000073659 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Gain of Function Mutation |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D057809 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | HEK293 Cells |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D006801 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Humans |
| mesh[6].qualifier_ui | Q000235 |
| mesh[6].descriptor_ui | D053565 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | genetics |
| mesh[6].descriptor_name | Hypercalciuria |
| mesh[7].qualifier_ui | Q000378 |
| mesh[7].descriptor_ui | D053565 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | metabolism |
| mesh[7].descriptor_name | Hypercalciuria |
| mesh[8].qualifier_ui | Q000235 |
| mesh[8].descriptor_ui | D006996 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | genetics |
| mesh[8].descriptor_name | Hypocalcemia |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D006996 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Hypocalcemia |
| mesh[10].qualifier_ui | Q000151 |
| mesh[10].descriptor_ui | D007011 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | congenital |
| mesh[10].descriptor_name | Hypoparathyroidism |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D007011 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | Hypoparathyroidism |
| mesh[12].qualifier_ui | Q000378 |
| mesh[12].descriptor_ui | D007011 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | metabolism |
| mesh[12].descriptor_name | Hypoparathyroidism |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D000073658 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Loss of Function Mutation |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D011487 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Protein Conformation |
| mesh[15].qualifier_ui | Q000235 |
| mesh[15].descriptor_ui | D044169 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | genetics |
| mesh[15].descriptor_name | Receptors, Calcium-Sensing |
| mesh[16].qualifier_ui | Q000378 |
| mesh[16].descriptor_ui | D044169 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | metabolism |
| mesh[16].descriptor_name | Receptors, Calcium-Sensing |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D016415 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Sequence Alignment |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D015398 |
| mesh[18].is_major_topic | True |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Signal Transduction |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D000595 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Amino Acid Sequence |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D020013 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Calcium Signaling |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D004252 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | DNA Mutational Analysis |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D000073659 |
| mesh[22].is_major_topic | True |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Gain of Function Mutation |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D057809 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | HEK293 Cells |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D006801 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Humans |
| mesh[25].qualifier_ui | Q000235 |
| mesh[25].descriptor_ui | D053565 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | genetics |
| mesh[25].descriptor_name | Hypercalciuria |
| mesh[26].qualifier_ui | Q000378 |
| mesh[26].descriptor_ui | D053565 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | metabolism |
| mesh[26].descriptor_name | Hypercalciuria |
| mesh[27].qualifier_ui | Q000235 |
| mesh[27].descriptor_ui | D006996 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | genetics |
| mesh[27].descriptor_name | Hypocalcemia |
| mesh[28].qualifier_ui | Q000378 |
| mesh[28].descriptor_ui | D006996 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | metabolism |
| mesh[28].descriptor_name | Hypocalcemia |
| mesh[29].qualifier_ui | Q000151 |
| mesh[29].descriptor_ui | D007011 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | congenital |
| mesh[29].descriptor_name | Hypoparathyroidism |
| mesh[30].qualifier_ui | Q000235 |
| mesh[30].descriptor_ui | D007011 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | genetics |
| mesh[30].descriptor_name | Hypoparathyroidism |
| mesh[31].qualifier_ui | Q000378 |
| mesh[31].descriptor_ui | D007011 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | metabolism |
| mesh[31].descriptor_name | Hypoparathyroidism |
| mesh[32].qualifier_ui | |
| mesh[32].descriptor_ui | D000073658 |
| mesh[32].is_major_topic | True |
| mesh[32].qualifier_name | |
| mesh[32].descriptor_name | Loss of Function Mutation |
| mesh[33].qualifier_ui | |
| mesh[33].descriptor_ui | D011487 |
| mesh[33].is_major_topic | False |
| mesh[33].qualifier_name | |
| mesh[33].descriptor_name | Protein Conformation |
| mesh[34].qualifier_ui | Q000235 |
| mesh[34].descriptor_ui | D044169 |
| mesh[34].is_major_topic | False |
| mesh[34].qualifier_name | genetics |
| mesh[34].descriptor_name | Receptors, Calcium-Sensing |
| mesh[35].qualifier_ui | Q000378 |
| mesh[35].descriptor_ui | D044169 |
| mesh[35].is_major_topic | False |
| mesh[35].qualifier_name | metabolism |
| mesh[35].descriptor_name | Receptors, Calcium-Sensing |
| mesh[36].qualifier_ui | |
| mesh[36].descriptor_ui | D016415 |
| mesh[36].is_major_topic | False |
| mesh[36].qualifier_name | |
| mesh[36].descriptor_name | Sequence Alignment |
| mesh[37].qualifier_ui | |
| mesh[37].descriptor_ui | D015398 |
| mesh[37].is_major_topic | True |
| mesh[37].qualifier_name | |
| mesh[37].descriptor_name | Signal Transduction |
| mesh[38].qualifier_ui | |
| mesh[38].descriptor_ui | D000595 |
| mesh[38].is_major_topic | False |
| mesh[38].qualifier_name | |
| mesh[38].descriptor_name | Amino Acid Sequence |
| mesh[39].qualifier_ui | |
| mesh[39].descriptor_ui | D020013 |
| mesh[39].is_major_topic | False |
| mesh[39].qualifier_name | |
| mesh[39].descriptor_name | Calcium Signaling |
| mesh[40].qualifier_ui | |
| mesh[40].descriptor_ui | D004252 |
| mesh[40].is_major_topic | False |
| mesh[40].qualifier_name | |
| mesh[40].descriptor_name | DNA Mutational Analysis |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D000073659 |
| mesh[41].is_major_topic | True |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | Gain of Function Mutation |
| mesh[42].qualifier_ui | |
| mesh[42].descriptor_ui | D057809 |
| mesh[42].is_major_topic | False |
| mesh[42].qualifier_name | |
| mesh[42].descriptor_name | HEK293 Cells |
| mesh[43].qualifier_ui | |
| mesh[43].descriptor_ui | D006801 |
| mesh[43].is_major_topic | False |
| mesh[43].qualifier_name | |
| mesh[43].descriptor_name | Humans |
| mesh[44].qualifier_ui | Q000235 |
| mesh[44].descriptor_ui | D053565 |
| mesh[44].is_major_topic | False |
| mesh[44].qualifier_name | genetics |
| mesh[44].descriptor_name | Hypercalciuria |
| mesh[45].qualifier_ui | Q000378 |
| mesh[45].descriptor_ui | D053565 |
| mesh[45].is_major_topic | False |
| mesh[45].qualifier_name | metabolism |
| mesh[45].descriptor_name | Hypercalciuria |
| mesh[46].qualifier_ui | Q000235 |
| mesh[46].descriptor_ui | D006996 |
| mesh[46].is_major_topic | False |
| mesh[46].qualifier_name | genetics |
| mesh[46].descriptor_name | Hypocalcemia |
| mesh[47].qualifier_ui | Q000378 |
| mesh[47].descriptor_ui | D006996 |
| mesh[47].is_major_topic | False |
| mesh[47].qualifier_name | metabolism |
| mesh[47].descriptor_name | Hypocalcemia |
| mesh[48].qualifier_ui | Q000151 |
| mesh[48].descriptor_ui | D007011 |
| mesh[48].is_major_topic | False |
| mesh[48].qualifier_name | congenital |
| mesh[48].descriptor_name | Hypoparathyroidism |
| mesh[49].qualifier_ui | Q000235 |
| mesh[49].descriptor_ui | D007011 |
| mesh[49].is_major_topic | False |
| mesh[49].qualifier_name | genetics |
| mesh[49].descriptor_name | Hypoparathyroidism |
| type | article |
| title | Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias |
| awards[0].id | https://openalex.org/G263527006 |
| awards[0].funder_id | https://openalex.org/F4320307874 |
| awards[0].display_name | |
| awards[0].funder_award_id | NF-SI-0514-10091 |
| awards[0].funder_display_name | Wellcome |
| awards[1].id | https://openalex.org/G2330666385 |
| awards[1].funder_id | https://openalex.org/F4320307846 |
| awards[1].display_name | |
| awards[1].funder_award_id | LT000021/2014-L |
| awards[1].funder_display_name | Human Frontier Science Program |
| awards[2].id | https://openalex.org/G6524872642 |
| awards[2].funder_id | https://openalex.org/F4320319985 |
| awards[2].display_name | |
| awards[2].funder_award_id | C20724/A14414 |
| awards[2].funder_display_name | Cancer Research UK |
| awards[3].id | https://openalex.org/G5821386806 |
| awards[3].funder_id | https://openalex.org/F4320307874 |
| awards[3].display_name | |
| awards[3].funder_award_id | 106995/Z/15/Z |
| awards[3].funder_display_name | Wellcome |
| awards[4].id | https://openalex.org/G8314207172 |
| awards[4].funder_id | https://openalex.org/F4320338335 |
| awards[4].display_name | Molecular analysis of the Hedgehog signal transduction complex in the primary cilium |
| awards[4].funder_award_id | 647278 |
| awards[4].funder_display_name | H2020 European Research Council |
| biblio.issue | 21 |
| biblio.volume | 27 |
| biblio.last_page | 3733 |
| biblio.first_page | 3720 |
| topics[0].id | https://openalex.org/T10169 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9997000098228455 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Protein Kinase Regulation and GTPase Signaling |
| topics[1].id | https://openalex.org/T12286 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9983000159263611 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2737 |
| topics[1].subfield.display_name | Physiology |
| topics[1].display_name | Erythrocyte Function and Pathophysiology |
| topics[2].id | https://openalex.org/T10493 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9968000054359436 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Ion channel regulation and function |
| funders[0].id | https://openalex.org/F4320307846 |
| funders[0].ror | https://ror.org/02ebx7v45 |
| funders[0].display_name | Human Frontier Science Program |
| funders[1].id | https://openalex.org/F4320307874 |
| funders[1].ror | https://ror.org/029chgv08 |
| funders[1].display_name | Wellcome |
| funders[2].id | https://openalex.org/F4320319985 |
| funders[2].ror | https://ror.org/054225q67 |
| funders[2].display_name | Cancer Research UK |
| funders[3].id | https://openalex.org/F4320319990 |
| funders[3].ror | https://ror.org/0187kwz08 |
| funders[3].display_name | National Institute for Health and Care Research |
| funders[4].id | https://openalex.org/F4320338335 |
| funders[4].ror | https://ror.org/0472cxd90 |
| funders[4].display_name | H2020 European Research Council |
| is_xpac | False |
| apc_list.value | 4181 |
| apc_list.currency | USD |
| apc_list.value_usd | 4181 |
| apc_paid.value | 4181 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 4181 |
| concepts[0].id | https://openalex.org/C142590548 |
| concepts[0].level | 4 |
| concepts[0].score | 0.8623851537704468 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q417801 |
| concepts[0].display_name | Calcium-sensing receptor |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.6974849700927734 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C28406088 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6532206535339355 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q5571762 |
| concepts[2].display_name | Extracellular |
| concepts[3].id | https://openalex.org/C11960822 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5974640846252441 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q242736 |
| concepts[3].display_name | Phosphorylation |
| concepts[4].id | https://openalex.org/C170493617 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5050652623176575 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[4].display_name | Receptor |
| concepts[5].id | https://openalex.org/C501734568 |
| concepts[5].level | 3 |
| concepts[5].score | 0.49271419644355774 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q42918 |
| concepts[5].display_name | Mutation |
| concepts[6].id | https://openalex.org/C57074206 |
| concepts[6].level | 3 |
| concepts[6].score | 0.48724165558815 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q14197552 |
| concepts[6].display_name | MAPK/ERK pathway |
| concepts[7].id | https://openalex.org/C62478195 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4804767668247223 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q828130 |
| concepts[7].display_name | Signal transduction |
| concepts[8].id | https://openalex.org/C95444343 |
| concepts[8].level | 1 |
| concepts[8].score | 0.4377394914627075 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[8].display_name | Cell biology |
| concepts[9].id | https://openalex.org/C135285700 |
| concepts[9].level | 3 |
| concepts[9].score | 0.4169888496398926 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q38173 |
| concepts[9].display_name | G protein-coupled receptor |
| concepts[10].id | https://openalex.org/C519063684 |
| concepts[10].level | 2 |
| concepts[10].score | 0.39454832673072815 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q706 |
| concepts[10].display_name | Calcium |
| concepts[11].id | https://openalex.org/C87975464 |
| concepts[11].level | 3 |
| concepts[11].score | 0.3437086343765259 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q910704 |
| concepts[11].display_name | Calcium metabolism |
| concepts[12].id | https://openalex.org/C55493867 |
| concepts[12].level | 1 |
| concepts[12].score | 0.29843926429748535 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[12].display_name | Biochemistry |
| concepts[13].id | https://openalex.org/C126322002 |
| concepts[13].level | 1 |
| concepts[13].score | 0.23931223154067993 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[13].display_name | Internal medicine |
| concepts[14].id | https://openalex.org/C104317684 |
| concepts[14].level | 2 |
| concepts[14].score | 0.1018034815788269 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[14].display_name | Gene |
| concepts[15].id | https://openalex.org/C71924100 |
| concepts[15].level | 0 |
| concepts[15].score | 0.06042414903640747 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[15].display_name | Medicine |
| keywords[0].id | https://openalex.org/keywords/calcium-sensing-receptor |
| keywords[0].score | 0.8623851537704468 |
| keywords[0].display_name | Calcium-sensing receptor |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.6974849700927734 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/extracellular |
| keywords[2].score | 0.6532206535339355 |
| keywords[2].display_name | Extracellular |
| keywords[3].id | https://openalex.org/keywords/phosphorylation |
| keywords[3].score | 0.5974640846252441 |
| keywords[3].display_name | Phosphorylation |
| keywords[4].id | https://openalex.org/keywords/receptor |
| keywords[4].score | 0.5050652623176575 |
| keywords[4].display_name | Receptor |
| keywords[5].id | https://openalex.org/keywords/mutation |
| keywords[5].score | 0.49271419644355774 |
| keywords[5].display_name | Mutation |
| keywords[6].id | https://openalex.org/keywords/mapk/erk-pathway |
| keywords[6].score | 0.48724165558815 |
| keywords[6].display_name | MAPK/ERK pathway |
| keywords[7].id | https://openalex.org/keywords/signal-transduction |
| keywords[7].score | 0.4804767668247223 |
| keywords[7].display_name | Signal transduction |
| keywords[8].id | https://openalex.org/keywords/cell-biology |
| keywords[8].score | 0.4377394914627075 |
| keywords[8].display_name | Cell biology |
| keywords[9].id | https://openalex.org/keywords/g-protein-coupled-receptor |
| keywords[9].score | 0.4169888496398926 |
| keywords[9].display_name | G protein-coupled receptor |
| keywords[10].id | https://openalex.org/keywords/calcium |
| keywords[10].score | 0.39454832673072815 |
| keywords[10].display_name | Calcium |
| keywords[11].id | https://openalex.org/keywords/calcium-metabolism |
| keywords[11].score | 0.3437086343765259 |
| keywords[11].display_name | Calcium metabolism |
| keywords[12].id | https://openalex.org/keywords/biochemistry |
| keywords[12].score | 0.29843926429748535 |
| keywords[12].display_name | Biochemistry |
| keywords[13].id | https://openalex.org/keywords/internal-medicine |
| keywords[13].score | 0.23931223154067993 |
| keywords[13].display_name | Internal medicine |
| keywords[14].id | https://openalex.org/keywords/gene |
| keywords[14].score | 0.1018034815788269 |
| keywords[14].display_name | Gene |
| keywords[15].id | https://openalex.org/keywords/medicine |
| keywords[15].score | 0.06042414903640747 |
| keywords[15].display_name | Medicine |
| language | en |
| locations[0].id | doi:10.1093/hmg/ddy263 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S166515463 |
| locations[0].source.issn | 0964-6906, 1460-2083 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0964-6906 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Human Molecular Genetics |
| locations[0].source.host_organization | https://openalex.org/P4310311648 |
| locations[0].source.host_organization_name | Oxford University Press |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| locations[0].source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Human Molecular Genetics |
| locations[0].landing_page_url | https://doi.org/10.1093/hmg/ddy263 |
| locations[1].id | pmid:30052933 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Human molecular genetics |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/30052933 |
| locations[2].id | pmh:oai:europepmc.org:5147336 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306400806 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Europe PMC (PubMed Central) |
| locations[2].source.host_organization | https://openalex.org/I1303153112 |
| locations[2].source.host_organization_name | European Bioinformatics Institute |
| locations[2].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[2].license | cc-by |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | http://europepmc.org/pmc/articles/PMC6196656 |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:6196656 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Hum Mol Genet |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/6196656 |
| locations[4].id | pmh:oai:pure.atira.dk:Publications/acc6a03f-2612-4086-a07c-6025d7e0bfcb |
| locations[4].is_oa | False |
| locations[4].source.id | https://openalex.org/S4306400216 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | Research Portal (King's College London) |
| locations[4].source.host_organization | https://openalex.org/I183935753 |
| locations[4].source.host_organization_name | King's College London |
| locations[4].source.host_organization_lineage | https://openalex.org/I183935753 |
| locations[4].license | |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | |
| locations[4].license_id | |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | |
| locations[4].landing_page_url | |
| locations[5].id | pmh:oai:pure.atira.dk:publications/acc6a03f-2612-4086-a07c-6025d7e0bfcb |
| locations[5].is_oa | False |
| locations[5].source.id | https://openalex.org/S4306400216 |
| locations[5].source.issn | |
| locations[5].source.type | repository |
| locations[5].source.is_oa | False |
| locations[5].source.issn_l | |
| locations[5].source.is_core | False |
| locations[5].source.is_in_doaj | False |
| locations[5].source.display_name | Research Portal (King's College London) |
| locations[5].source.host_organization | https://openalex.org/I183935753 |
| locations[5].source.host_organization_name | King's College London |
| locations[5].source.host_organization_lineage | https://openalex.org/I183935753 |
| locations[5].license | |
| locations[5].pdf_url | |
| locations[5].version | submittedVersion |
| locations[5].raw_type | |
| locations[5].license_id | |
| locations[5].is_accepted | False |
| locations[5].is_published | False |
| locations[5].raw_source_name | |
| locations[5].landing_page_url | https://research.birmingham.ac.uk/portal/en/publications/calciumsensing-receptor-residues-with-loss-and-gainoffunction-mutations-are-located-in-regions-of-conformational-change-and-cause-signalling-bias(acc6a03f-2612-4086-a07c-6025d7e0bfcb).html |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5011297390 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1361-9174 |
| authorships[0].author.display_name | Caroline M. Gorvin |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I40120149, https://openalex.org/I4210129944 |
| authorships[0].affiliations[0].raw_affiliation_string | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK |
| authorships[0].institutions[0].id | https://openalex.org/I4210129944 |
| authorships[0].institutions[0].ror | https://ror.org/03myafa32 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I40120149, https://openalex.org/I4210129944, https://openalex.org/I4210147381 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | Oxford Centre for Diabetes, Endocrinology and Metabolism |
| authorships[0].institutions[1].id | https://openalex.org/I40120149 |
| authorships[0].institutions[1].ror | https://ror.org/052gg0110 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I40120149 |
| authorships[0].institutions[1].country_code | GB |
| authorships[0].institutions[1].display_name | University of Oxford |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Caroline M Gorvin |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK |
| authorships[1].author.id | https://openalex.org/A5112892810 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Morten Frost |
| authorships[1].countries | DK, GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I40120149, https://openalex.org/I4210129944 |
| authorships[1].affiliations[0].raw_affiliation_string | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I177969490 |
| authorships[1].affiliations[1].raw_affiliation_string | University of Southern Denmark, Odense C, Denmark |
| authorships[1].institutions[0].id | https://openalex.org/I177969490 |
| authorships[1].institutions[0].ror | https://ror.org/03yrrjy16 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I177969490 |
| authorships[1].institutions[0].country_code | DK |
| authorships[1].institutions[0].display_name | University of Southern Denmark |
| authorships[1].institutions[1].id | https://openalex.org/I4210129944 |
| authorships[1].institutions[1].ror | https://ror.org/03myafa32 |
| authorships[1].institutions[1].type | facility |
| authorships[1].institutions[1].lineage | https://openalex.org/I40120149, https://openalex.org/I4210129944, https://openalex.org/I4210147381 |
| authorships[1].institutions[1].country_code | GB |
| authorships[1].institutions[1].display_name | Oxford Centre for Diabetes, Endocrinology and Metabolism |
| authorships[1].institutions[2].id | https://openalex.org/I40120149 |
| authorships[1].institutions[2].ror | https://ror.org/052gg0110 |
| authorships[1].institutions[2].type | education |
| authorships[1].institutions[2].lineage | https://openalex.org/I40120149 |
| authorships[1].institutions[2].country_code | GB |
| authorships[1].institutions[2].display_name | University of Oxford |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Morten Frost |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK, University of Southern Denmark, Odense C, Denmark |
| authorships[2].author.id | https://openalex.org/A5016481779 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-4847-5529 |
| authorships[2].author.display_name | Tomas Malinauskas |
| authorships[2].countries | GB |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I1336263701, https://openalex.org/I40120149 |
| authorships[2].affiliations[0].raw_affiliation_string | Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK |
| authorships[2].institutions[0].id | https://openalex.org/I1336263701 |
| authorships[2].institutions[0].ror | https://ror.org/01rjnta51 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I1336263701, https://openalex.org/I40120149 |
| authorships[2].institutions[0].country_code | GB |
| authorships[2].institutions[0].display_name | Centre for Human Genetics |
| authorships[2].institutions[1].id | https://openalex.org/I40120149 |
| authorships[2].institutions[1].ror | https://ror.org/052gg0110 |
| authorships[2].institutions[1].type | education |
| authorships[2].institutions[1].lineage | https://openalex.org/I40120149 |
| authorships[2].institutions[1].country_code | GB |
| authorships[2].institutions[1].display_name | University of Oxford |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Tomas Malinauskas |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK |
| authorships[3].author.id | https://openalex.org/A5111597545 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Treena Cranston |
| authorships[3].countries | GB |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I2801179840 |
| authorships[3].affiliations[0].raw_affiliation_string | Oxford Molecular Genetics Laboratory, Churchill Hospital, Oxford OX3 7LJ, UK |
| authorships[3].institutions[0].id | https://openalex.org/I2801179840 |
| authorships[3].institutions[0].ror | https://ror.org/009vheq40 |
| authorships[3].institutions[0].type | healthcare |
| authorships[3].institutions[0].lineage | https://openalex.org/I2801179840, https://openalex.org/I4210147381 |
| authorships[3].institutions[0].country_code | GB |
| authorships[3].institutions[0].display_name | Churchill Hospital |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Treena Cranston |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Oxford Molecular Genetics Laboratory, Churchill Hospital, Oxford OX3 7LJ, UK |
| authorships[4].author.id | https://openalex.org/A5033507754 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Hannah Boon |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I2801179840 |
| authorships[4].affiliations[0].raw_affiliation_string | Oxford Molecular Genetics Laboratory, Churchill Hospital, Oxford OX3 7LJ, UK |
| authorships[4].institutions[0].id | https://openalex.org/I2801179840 |
| authorships[4].institutions[0].ror | https://ror.org/009vheq40 |
| authorships[4].institutions[0].type | healthcare |
| authorships[4].institutions[0].lineage | https://openalex.org/I2801179840, https://openalex.org/I4210147381 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | Churchill Hospital |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Hannah Boon |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Oxford Molecular Genetics Laboratory, Churchill Hospital, Oxford OX3 7LJ, UK |
| authorships[5].author.id | https://openalex.org/A5037626610 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-6635-3621 |
| authorships[5].author.display_name | Christian Siebold |
| authorships[5].countries | GB |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I1336263701, https://openalex.org/I40120149 |
| authorships[5].affiliations[0].raw_affiliation_string | Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK |
| authorships[5].institutions[0].id | https://openalex.org/I1336263701 |
| authorships[5].institutions[0].ror | https://ror.org/01rjnta51 |
| authorships[5].institutions[0].type | facility |
| authorships[5].institutions[0].lineage | https://openalex.org/I1336263701, https://openalex.org/I40120149 |
| authorships[5].institutions[0].country_code | GB |
| authorships[5].institutions[0].display_name | Centre for Human Genetics |
| authorships[5].institutions[1].id | https://openalex.org/I40120149 |
| authorships[5].institutions[1].ror | https://ror.org/052gg0110 |
| authorships[5].institutions[1].type | education |
| authorships[5].institutions[1].lineage | https://openalex.org/I40120149 |
| authorships[5].institutions[1].country_code | GB |
| authorships[5].institutions[1].display_name | University of Oxford |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Christian Siebold |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK |
| authorships[6].author.id | https://openalex.org/A5016293875 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-3834-1893 |
| authorships[6].author.display_name | E. Yvonne Jones |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1336263701, https://openalex.org/I40120149 |
| authorships[6].affiliations[0].raw_affiliation_string | Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK |
| authorships[6].institutions[0].id | https://openalex.org/I1336263701 |
| authorships[6].institutions[0].ror | https://ror.org/01rjnta51 |
| authorships[6].institutions[0].type | facility |
| authorships[6].institutions[0].lineage | https://openalex.org/I1336263701, https://openalex.org/I40120149 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | Centre for Human Genetics |
| authorships[6].institutions[1].id | https://openalex.org/I40120149 |
| authorships[6].institutions[1].ror | https://ror.org/052gg0110 |
| authorships[6].institutions[1].type | education |
| authorships[6].institutions[1].lineage | https://openalex.org/I40120149 |
| authorships[6].institutions[1].country_code | GB |
| authorships[6].institutions[1].display_name | University of Oxford |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | E Yvonne Jones |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK |
| authorships[7].author.id | https://openalex.org/A5063333149 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-2975-5170 |
| authorships[7].author.display_name | Fadil Hannan |
| authorships[7].countries | GB |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I40120149, https://openalex.org/I4210129944 |
| authorships[7].affiliations[0].raw_affiliation_string | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I146655781 |
| authorships[7].affiliations[1].raw_affiliation_string | Institute of Ageing and Chronic Disease, University of Liverpool, Liverpool, UK |
| authorships[7].institutions[0].id | https://openalex.org/I4210129944 |
| authorships[7].institutions[0].ror | https://ror.org/03myafa32 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I40120149, https://openalex.org/I4210129944, https://openalex.org/I4210147381 |
| authorships[7].institutions[0].country_code | GB |
| authorships[7].institutions[0].display_name | Oxford Centre for Diabetes, Endocrinology and Metabolism |
| authorships[7].institutions[1].id | https://openalex.org/I146655781 |
| authorships[7].institutions[1].ror | https://ror.org/04xs57h96 |
| authorships[7].institutions[1].type | education |
| authorships[7].institutions[1].lineage | https://openalex.org/I146655781 |
| authorships[7].institutions[1].country_code | GB |
| authorships[7].institutions[1].display_name | University of Liverpool |
| authorships[7].institutions[2].id | https://openalex.org/I40120149 |
| authorships[7].institutions[2].ror | https://ror.org/052gg0110 |
| authorships[7].institutions[2].type | education |
| authorships[7].institutions[2].lineage | https://openalex.org/I40120149 |
| authorships[7].institutions[2].country_code | GB |
| authorships[7].institutions[2].display_name | University of Oxford |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Fadil M Hannan |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK, Institute of Ageing and Chronic Disease, University of Liverpool, Liverpool, UK |
| authorships[8].author.id | https://openalex.org/A5080589561 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-1438-3220 |
| authorships[8].author.display_name | Rajesh V. Thakker |
| authorships[8].countries | GB |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I40120149, https://openalex.org/I4210129944 |
| authorships[8].affiliations[0].raw_affiliation_string | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK |
| authorships[8].institutions[0].id | https://openalex.org/I4210129944 |
| authorships[8].institutions[0].ror | https://ror.org/03myafa32 |
| authorships[8].institutions[0].type | facility |
| authorships[8].institutions[0].lineage | https://openalex.org/I40120149, https://openalex.org/I4210129944, https://openalex.org/I4210147381 |
| authorships[8].institutions[0].country_code | GB |
| authorships[8].institutions[0].display_name | Oxford Centre for Diabetes, Endocrinology and Metabolism |
| authorships[8].institutions[1].id | https://openalex.org/I40120149 |
| authorships[8].institutions[1].ror | https://ror.org/052gg0110 |
| authorships[8].institutions[1].type | education |
| authorships[8].institutions[1].lineage | https://openalex.org/I40120149 |
| authorships[8].institutions[1].country_code | GB |
| authorships[8].institutions[1].display_name | University of Oxford |
| authorships[8].author_position | last |
| authorships[8].raw_author_name | Rajesh V Thakker |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | Academic Endocrine Unit, Radcliffe Department of Medicine, Oxford Centre for Diabetes, Endocrinology and Metabolism (OCDEM), University of Oxford, Oxford OX3 7LJ, UK |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdf |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10169 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9997000098228455 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Protein Kinase Regulation and GTPase Signaling |
| related_works | https://openalex.org/W2599174518, https://openalex.org/W2035469386, https://openalex.org/W2984175546, https://openalex.org/W2563976934, https://openalex.org/W2488726591, https://openalex.org/W1996946402, https://openalex.org/W91001492, https://openalex.org/W118511553, https://openalex.org/W2018887905, https://openalex.org/W2990201100 |
| cited_by_count | 28 |
| counts_by_year[0].year | 2024 |
| counts_by_year[0].cited_by_count | 5 |
| counts_by_year[1].year | 2023 |
| counts_by_year[1].cited_by_count | 2 |
| counts_by_year[2].year | 2022 |
| counts_by_year[2].cited_by_count | 2 |
| counts_by_year[3].year | 2021 |
| counts_by_year[3].cited_by_count | 7 |
| counts_by_year[4].year | 2020 |
| counts_by_year[4].cited_by_count | 7 |
| counts_by_year[5].year | 2019 |
| counts_by_year[5].cited_by_count | 3 |
| counts_by_year[6].year | 2018 |
| counts_by_year[6].cited_by_count | 2 |
| locations_count | 6 |
| best_oa_location.id | doi:10.1093/hmg/ddy263 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S166515463 |
| best_oa_location.source.issn | 0964-6906, 1460-2083 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0964-6906 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Human Molecular Genetics |
| best_oa_location.source.host_organization | https://openalex.org/P4310311648 |
| best_oa_location.source.host_organization_name | Oxford University Press |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| best_oa_location.source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Human Molecular Genetics |
| best_oa_location.landing_page_url | https://doi.org/10.1093/hmg/ddy263 |
| primary_location.id | doi:10.1093/hmg/ddy263 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S166515463 |
| primary_location.source.issn | 0964-6906, 1460-2083 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0964-6906 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Human Molecular Genetics |
| primary_location.source.host_organization | https://openalex.org/P4310311648 |
| primary_location.source.host_organization_name | Oxford University Press |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| primary_location.source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://academic.oup.com/hmg/article-pdf/27/21/3720/26127123/ddy263.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Human Molecular Genetics |
| primary_location.landing_page_url | https://doi.org/10.1093/hmg/ddy263 |
| publication_date | 2018-07-19 |
| publication_year | 2018 |
| referenced_works | https://openalex.org/W1973914285, https://openalex.org/W1973346467, https://openalex.org/W2485780168, https://openalex.org/W2408576125, https://openalex.org/W1998850010, https://openalex.org/W2162834665, https://openalex.org/W1549316628, https://openalex.org/W2183475472, https://openalex.org/W2008810242, https://openalex.org/W2788427401, https://openalex.org/W2522709216, https://openalex.org/W1984875786, https://openalex.org/W2065678231, https://openalex.org/W2171810658, https://openalex.org/W2126653660, https://openalex.org/W2107900599, https://openalex.org/W2139736883, https://openalex.org/W2083472004, https://openalex.org/W1986470122, https://openalex.org/W2157138210, https://openalex.org/W2105364782, https://openalex.org/W2256016639, https://openalex.org/W20573415, https://openalex.org/W4255760236, https://openalex.org/W2014979626, https://openalex.org/W2315593339, https://openalex.org/W2080448298, https://openalex.org/W2057602534, https://openalex.org/W2018897042, https://openalex.org/W2008728387, https://openalex.org/W2034941411, https://openalex.org/W2052091241, https://openalex.org/W2087163465, https://openalex.org/W2071539007, https://openalex.org/W2167857702, https://openalex.org/W2607506319, https://openalex.org/W2618233912, https://openalex.org/W1989969172, https://openalex.org/W2084401477, https://openalex.org/W2093640268, https://openalex.org/W2077553389, https://openalex.org/W1601065938, https://openalex.org/W2111373319, https://openalex.org/W2023287752, https://openalex.org/W1851767352, https://openalex.org/W2122732537, https://openalex.org/W2127322768, https://openalex.org/W2082667898, https://openalex.org/W6808128415, https://openalex.org/W2587753722, https://openalex.org/W2054481493, https://openalex.org/W2750372408, https://openalex.org/W2580073630, https://openalex.org/W2399759053, https://openalex.org/W1607931568, https://openalex.org/W2036145275, https://openalex.org/W4211196614, https://openalex.org/W4211156111 |
| referenced_works_count | 58 |
| abstract_inverted_index.a | 5, 239 |
| abstract_inverted_index.An | 108 |
| abstract_inverted_index.In | 168 |
| abstract_inverted_index.an | 170 |
| abstract_inverted_index.as | 99, 214 |
| abstract_inverted_index.at | 206, 231 |
| abstract_inverted_index.be | 204 |
| abstract_inverted_index.by | 42, 133, 176 |
| abstract_inverted_index.in | 36, 114, 241 |
| abstract_inverted_index.is | 4 |
| abstract_inverted_index.of | 18, 33, 45, 91, 110, 188, 210 |
| abstract_inverted_index.or | 47, 58, 74 |
| abstract_inverted_index.to | 24, 53, 102, 148, 178, 203 |
| abstract_inverted_index.we | 81 |
| abstract_inverted_index.1/2 | 22 |
| abstract_inverted_index.ADH | 136 |
| abstract_inverted_index.ERK | 75, 166, 184 |
| abstract_inverted_index.FHH | 134 |
| abstract_inverted_index.For | 153 |
| abstract_inverted_index.The | 0, 30 |
| abstract_inverted_index.act | 98 |
| abstract_inverted_index.and | 16, 80, 94, 117, 128, 135, 159, 219, 237 |
| abstract_inverted_index.are | 88, 131, 229 |
| abstract_inverted_index.for | 234 |
| abstract_inverted_index.has | 39 |
| abstract_inverted_index.may | 97 |
| abstract_inverted_index.not | 77 |
| abstract_inverted_index.our | 223 |
| abstract_inverted_index.the | 34, 43, 65, 69, 89, 211, 215 |
| abstract_inverted_index.two | 155 |
| abstract_inverted_index.via | 11, 72 |
| abstract_inverted_index.>300 | 115 |
| abstract_inverted_index.CaSR | 35, 49, 70, 85, 111, 191, 212, 235 |
| abstract_inverted_index.been | 40, 78 |
| abstract_inverted_index.both | 92 |
| abstract_inverted_index.five | 121, 190 |
| abstract_inverted_index.four | 196 |
| abstract_inverted_index.have | 76 |
| abstract_inverted_index.lead | 52 |
| abstract_inverted_index.play | 238 |
| abstract_inverted_index.role | 240 |
| abstract_inverted_index.site | 90 |
| abstract_inverted_index.some | 84 |
| abstract_inverted_index.such | 213 |
| abstract_inverted_index.that | 9, 51, 83, 130, 226 |
| abstract_inverted_index.with | 195 |
| abstract_inverted_index.(ERK) | 23 |
| abstract_inverted_index.(FHH) | 57 |
| abstract_inverted_index.Ca2+e | 37 |
| abstract_inverted_index.Ca2+i | 73, 162, 180 |
| abstract_inverted_index.Thus, | 222 |
| abstract_inverted_index.bias. | 152, 244 |
| abstract_inverted_index.cells | 143 |
| abstract_inverted_index.dimer | 217 |
| abstract_inverted_index.loss- | 46, 93 |
| abstract_inverted_index.sites | 232 |
| abstract_inverted_index.these | 106, 189, 201 |
| abstract_inverted_index.using | 141 |
| abstract_inverted_index.which | 87 |
| abstract_inverted_index.while | 182 |
| abstract_inverted_index.(ADH), | 62 |
| abstract_inverted_index.(CaSR) | 3 |
| abstract_inverted_index.HEK293 | 142 |
| abstract_inverted_index.Ser820 | 127 |
| abstract_inverted_index.active | 208 |
| abstract_inverted_index.direct | 103 |
| abstract_inverted_index.kinase | 21 |
| abstract_inverted_index.showed | 144 |
| abstract_inverted_index.(Ca2+e) | 28 |
| abstract_inverted_index.(Ca2+i) | 14 |
| abstract_inverted_index.(Gln27, | 124 |
| abstract_inverted_index.Asn178, | 125 |
| abstract_inverted_index.Ser657, | 126 |
| abstract_inverted_index.Thr828) | 129 |
| abstract_inverted_index.calcium | 13, 27 |
| abstract_inverted_index.central | 31 |
| abstract_inverted_index.display | 150 |
| abstract_inverted_index.domain. | 221 |
| abstract_inverted_index.leading | 177 |
| abstract_inverted_index.located | 205, 230 |
| abstract_inverted_index.regions | 209 |
| abstract_inverted_index.residue | 146 |
| abstract_inverted_index.signals | 10, 71 |
| abstract_inverted_index.studies | 140 |
| abstract_inverted_index.through | 105 |
| abstract_inverted_index.whether | 68 |
| abstract_inverted_index.without | 164 |
| abstract_inverted_index.However, | 64 |
| abstract_inverted_index.affected | 132 |
| abstract_inverted_index.altering | 165 |
| abstract_inverted_index.analysis | 109, 187 |
| abstract_inverted_index.commonly | 149 |
| abstract_inverted_index.critical | 233 |
| abstract_inverted_index.dominant | 60 |
| abstract_inverted_index.example, | 154 |
| abstract_inverted_index.familial | 54 |
| abstract_inverted_index.findings | 224 |
| abstract_inverted_index.impaired | 161 |
| abstract_inverted_index.indicate | 225 |
| abstract_inverted_index.mutation | 173 |
| abstract_inverted_index.probands | 119 |
| abstract_inverted_index.receptor | 2, 8 |
| abstract_inverted_index.regulate | 25 |
| abstract_inverted_index.reported | 197 |
| abstract_inverted_index.residues | 123, 193, 199, 202, 228 |
| abstract_inverted_index.revealed | 120, 200 |
| abstract_inverted_index.switches | 101 |
| abstract_inverted_index.together | 194 |
| abstract_inverted_index.autosomal | 59 |
| abstract_inverted_index.contrast, | 169 |
| abstract_inverted_index.increased | 179 |
| abstract_inverted_index.interface | 218 |
| abstract_inverted_index.mediating | 242 |
| abstract_inverted_index.molecular | 100 |
| abstract_inverted_index.mutations | 50, 112, 147, 157 |
| abstract_inverted_index.pathways. | 107 |
| abstract_inverted_index.residues, | 86 |
| abstract_inverted_index.uncoupled | 174 |
| abstract_inverted_index.Functional | 138 |
| abstract_inverted_index.Structural | 186 |
| abstract_inverted_index.activation | 236 |
| abstract_inverted_index.decreasing | 183 |
| abstract_inverted_index.expression | 139 |
| abstract_inverted_index.identified | 113 |
| abstract_inverted_index.importance | 32 |
| abstract_inverted_index.mechanisms | 66 |
| abstract_inverted_index.mutations, | 96 |
| abstract_inverted_index.mutations. | 137 |
| abstract_inverted_index.signalling | 104, 151, 163, 175, 243 |
| abstract_inverted_index.determining | 67 |
| abstract_inverted_index.homeostasis | 38 |
| abstract_inverted_index.homodimeric | 6 |
| abstract_inverted_index.p.Ser657Cys | 172 |
| abstract_inverted_index.(p.Asn178Asp | 158 |
| abstract_inverted_index.demonstrated | 41 |
| abstract_inverted_index.established, | 79 |
| abstract_inverted_index.homeostasis. | 29 |
| abstract_inverted_index.hypothesised | 82 |
| abstract_inverted_index.mobilisation | 15 |
| abstract_inverted_index.mobilization | 181 |
| abstract_inverted_index.p.Ser820Ala) | 160 |
| abstract_inverted_index.extracellular | 19, 26, 216 |
| abstract_inverted_index.hypocalcaemia | 61 |
| abstract_inverted_index.hypocalcaemic | 118 |
| abstract_inverted_index.hypocalciuric | 55 |
| abstract_inverted_index.intracellular | 12 |
| abstract_inverted_index.respectively. | 63 |
| abstract_inverted_index.transmembrane | 220 |
| abstract_inverted_index.ADH-associated | 171 |
| abstract_inverted_index.FHH-associated | 156 |
| abstract_inverted_index.disease-switch | 145, 192, 198, 227 |
| abstract_inverted_index.hypercalcaemia | 56 |
| abstract_inverted_index.hypercalcaemic | 116 |
| abstract_inverted_index.identification | 44 |
| abstract_inverted_index.calcium-sensing | 1 |
| abstract_inverted_index.phosphorylation | 17 |
| abstract_inverted_index.'disease-switch' | 122 |
| abstract_inverted_index.conformationally | 207 |
| abstract_inverted_index.gain-of-function | 48, 95 |
| abstract_inverted_index.phosphorylation. | 167, 185 |
| abstract_inverted_index.signal-regulated | 20 |
| abstract_inverted_index.G-protein-coupled | 7 |
| cited_by_percentile_year.max | 99 |
| cited_by_percentile_year.min | 94 |
| corresponding_author_ids | https://openalex.org/A5080589561 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 9 |
| corresponding_institution_ids | https://openalex.org/I40120149, https://openalex.org/I4210129944 |
| citation_normalized_percentile.value | 0.85673539 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |