Challenges and progress in interpretation of non-coding genetic variants associated with human disease Article Swipe
 YOU?
·
· 2017
· Open Access
·
· DOI: https://doi.org/10.1177/1535370217713750
YOU?
·
· 2017
· Open Access
·
· DOI: https://doi.org/10.1177/1535370217713750
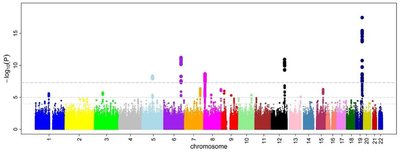
Genome-wide association studies have shown that the far majority of disease-associated variants reside in the non-coding regions of the genome, suggesting that gene regulatory changes contribute to disease risk. To identify truly causal non-coding variants and their affected target genes remains challenging but is a critical step to translate the genetic associations to molecular mechanisms and ultimately clinical applications. Here we review genomic/epigenomic resources and in silico tools that can be used to identify causal non-coding variants and experimental strategies to validate their functionalities. Impact statement Most signals from genome-wide association studies (GWASs) map to the non-coding genome, and functional interpretation of these associations remained challenging. We reviewed recent progress in methodologies of studying the non-coding genome and argued that no single approach allows one to effectively identify the causal regulatory variants from GWAS results. By illustrating the advantages and limitations of each method, our review potentially provided a guideline for taking a combinatorial approach to accurately predict, prioritize, and eventually experimentally validate the causal variants.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1177/1535370217713750
- OA Status
- green
- Cited By
- 70
- References
- 97
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2621435244
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2621435244Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1177/1535370217713750Digital Object Identifier
- Title
-
Challenges and progress in interpretation of non-coding genetic variants associated with human diseaseWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2017Year of publication
- Publication date
-
2017-06-05Full publication date if available
- Authors
-
Yizhou Zhu, Cagdas Tazearslan, Yousin SuhList of authors in order
- Landing page
-
https://doi.org/10.1177/1535370217713750Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.ncbi.nlm.nih.gov/pmc/articles/5529005Direct OA link when available
- Concepts
-
Genome-wide association study, Computational biology, Genetic association, Epigenomics, Genome, Human genome, Genomics, Biology, Computer science, Genetics, Gene, Single-nucleotide polymorphism, Genotype, DNA methylation, Gene expressionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
70Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 4, 2024: 22, 2023: 11, 2022: 4, 2021: 6Per-year citation counts (last 5 years)
- References (count)
-
97Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2621435244 |
|---|---|
| doi | https://doi.org/10.1177/1535370217713750 |
| ids.doi | https://doi.org/10.1177/1535370217713750 |
| ids.mag | 2621435244 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/28581336 |
| ids.openalex | https://openalex.org/W2621435244 |
| fwci | 3.07313832 |
| mesh[0].qualifier_ui | Q000379 |
| mesh[0].descriptor_ui | D019295 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | methods |
| mesh[0].descriptor_name | Computational Biology |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D044127 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Epigenesis, Genetic |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D020022 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Genetic Predisposition to Disease |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D055106 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Genome-Wide Association Study |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D006801 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Humans |
| mesh[5].qualifier_ui | Q000235 |
| mesh[5].descriptor_ui | D012045 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | genetics |
| mesh[5].descriptor_name | Regulatory Sequences, Nucleic Acid |
| mesh[6].qualifier_ui | Q000379 |
| mesh[6].descriptor_ui | D019295 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | methods |
| mesh[6].descriptor_name | Computational Biology |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D044127 |
| mesh[7].is_major_topic | True |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Epigenesis, Genetic |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D020022 |
| mesh[8].is_major_topic | True |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Genetic Predisposition to Disease |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D055106 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Genome-Wide Association Study |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D006801 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Humans |
| mesh[11].qualifier_ui | Q000235 |
| mesh[11].descriptor_ui | D012045 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | genetics |
| mesh[11].descriptor_name | Regulatory Sequences, Nucleic Acid |
| type | article |
| title | Challenges and progress in interpretation of non-coding genetic variants associated with human disease |
| biblio.issue | 13 |
| biblio.volume | 242 |
| biblio.last_page | 1334 |
| biblio.first_page | 1325 |
| topics[0].id | https://openalex.org/T10222 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9991000294685364 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Genomics and Chromatin Dynamics |
| topics[1].id | https://openalex.org/T11213 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9976999759674072 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genomic variations and chromosomal abnormalities |
| topics[2].id | https://openalex.org/T10261 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9970999956130981 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genetic Associations and Epidemiology |
| funders[0].id | https://openalex.org/F4320332161 |
| funders[0].ror | https://ror.org/01cwqze88 |
| funders[0].display_name | National Institutes of Health |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7002468109130859 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C70721500 |
| concepts[1].level | 1 |
| concepts[1].score | 0.6095942854881287 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[1].display_name | Computational biology |
| concepts[2].id | https://openalex.org/C186413461 |
| concepts[2].level | 5 |
| concepts[2].score | 0.5980985760688782 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[2].display_name | Genetic association |
| concepts[3].id | https://openalex.org/C121912465 |
| concepts[3].level | 5 |
| concepts[3].score | 0.5910440683364868 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q3589153 |
| concepts[3].display_name | Epigenomics |
| concepts[4].id | https://openalex.org/C141231307 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5257723331451416 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[4].display_name | Genome |
| concepts[5].id | https://openalex.org/C197077220 |
| concepts[5].level | 4 |
| concepts[5].score | 0.478641152381897 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q720988 |
| concepts[5].display_name | Human genome |
| concepts[6].id | https://openalex.org/C189206191 |
| concepts[6].level | 4 |
| concepts[6].score | 0.43876099586486816 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q222046 |
| concepts[6].display_name | Genomics |
| concepts[7].id | https://openalex.org/C86803240 |
| concepts[7].level | 0 |
| concepts[7].score | 0.4088508188724518 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[7].display_name | Biology |
| concepts[8].id | https://openalex.org/C41008148 |
| concepts[8].level | 0 |
| concepts[8].score | 0.35872769355773926 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[8].display_name | Computer science |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.33541208505630493 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C104317684 |
| concepts[10].level | 2 |
| concepts[10].score | 0.3080699145793915 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[10].display_name | Gene |
| concepts[11].id | https://openalex.org/C153209595 |
| concepts[11].level | 4 |
| concepts[11].score | 0.19386476278305054 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[11].display_name | Single-nucleotide polymorphism |
| concepts[12].id | https://openalex.org/C135763542 |
| concepts[12].level | 3 |
| concepts[12].score | 0.0704655647277832 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[12].display_name | Genotype |
| concepts[13].id | https://openalex.org/C190727270 |
| concepts[13].level | 4 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q874745 |
| concepts[13].display_name | DNA methylation |
| concepts[14].id | https://openalex.org/C150194340 |
| concepts[14].level | 3 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[14].display_name | Gene expression |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.7002468109130859 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/computational-biology |
| keywords[1].score | 0.6095942854881287 |
| keywords[1].display_name | Computational biology |
| keywords[2].id | https://openalex.org/keywords/genetic-association |
| keywords[2].score | 0.5980985760688782 |
| keywords[2].display_name | Genetic association |
| keywords[3].id | https://openalex.org/keywords/epigenomics |
| keywords[3].score | 0.5910440683364868 |
| keywords[3].display_name | Epigenomics |
| keywords[4].id | https://openalex.org/keywords/genome |
| keywords[4].score | 0.5257723331451416 |
| keywords[4].display_name | Genome |
| keywords[5].id | https://openalex.org/keywords/human-genome |
| keywords[5].score | 0.478641152381897 |
| keywords[5].display_name | Human genome |
| keywords[6].id | https://openalex.org/keywords/genomics |
| keywords[6].score | 0.43876099586486816 |
| keywords[6].display_name | Genomics |
| keywords[7].id | https://openalex.org/keywords/biology |
| keywords[7].score | 0.4088508188724518 |
| keywords[7].display_name | Biology |
| keywords[8].id | https://openalex.org/keywords/computer-science |
| keywords[8].score | 0.35872769355773926 |
| keywords[8].display_name | Computer science |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.33541208505630493 |
| keywords[9].display_name | Genetics |
| keywords[10].id | https://openalex.org/keywords/gene |
| keywords[10].score | 0.3080699145793915 |
| keywords[10].display_name | Gene |
| keywords[11].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[11].score | 0.19386476278305054 |
| keywords[11].display_name | Single-nucleotide polymorphism |
| keywords[12].id | https://openalex.org/keywords/genotype |
| keywords[12].score | 0.0704655647277832 |
| keywords[12].display_name | Genotype |
| language | en |
| locations[0].id | doi:10.1177/1535370217713750 |
| locations[0].is_oa | False |
| locations[0].source.id | https://openalex.org/S67514239 |
| locations[0].source.issn | 1535-3699, 1535-3702 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 1535-3699 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Experimental Biology and Medicine |
| locations[0].source.host_organization | https://openalex.org/P4310320017 |
| locations[0].source.host_organization_name | SAGE Publishing |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320017 |
| locations[0].source.host_organization_lineage_names | SAGE Publishing |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Experimental Biology and Medicine |
| locations[0].landing_page_url | https://doi.org/10.1177/1535370217713750 |
| locations[1].id | pmid:28581336 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Experimental biology and medicine (Maywood, N.J.) |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/28581336 |
| locations[2].id | pmh:oai:europepmc.org:4659878 |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306400806 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | Europe PMC (PubMed Central) |
| locations[2].source.host_organization | https://openalex.org/I1303153112 |
| locations[2].source.host_organization_name | European Bioinformatics Institute |
| locations[2].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | http://europepmc.org/pmc/articles/PMC5529005 |
| locations[3].id | pmh:oai:pubmedcentral.nih.gov:5529005 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S2764455111 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | PubMed Central |
| locations[3].source.host_organization | https://openalex.org/I1299303238 |
| locations[3].source.host_organization_name | National Institutes of Health |
| locations[3].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Exp Biol Med (Maywood) |
| locations[3].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/5529005 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5016512840 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-0234-4603 |
| authorships[0].author.display_name | Yizhou Zhu |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I129975664 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[0].institutions[0].id | https://openalex.org/I129975664 |
| authorships[0].institutions[0].ror | https://ror.org/05cf8a891 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I129975664, https://openalex.org/I4210112371 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Albert Einstein College of Medicine |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Yizhou Zhu |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[1].author.id | https://openalex.org/A5067847561 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Cagdas Tazearslan |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I129975664 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[1].institutions[0].id | https://openalex.org/I129975664 |
| authorships[1].institutions[0].ror | https://ror.org/05cf8a891 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I129975664, https://openalex.org/I4210112371 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Albert Einstein College of Medicine |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Cagdas Tazearslan |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[2].author.id | https://openalex.org/A5080863597 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-6014-3084 |
| authorships[2].author.display_name | Yousin Suh |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I129975664 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Ophthalmology & Visual Sciences, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I129975664 |
| authorships[2].affiliations[1].raw_affiliation_string | Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[2].affiliations[2].institution_ids | https://openalex.org/I129975664 |
| authorships[2].affiliations[2].raw_affiliation_string | Department of Medicine, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| authorships[2].institutions[0].id | https://openalex.org/I129975664 |
| authorships[2].institutions[0].ror | https://ror.org/05cf8a891 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I129975664, https://openalex.org/I4210112371 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Albert Einstein College of Medicine |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Yousin Suh |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA, Department of Medicine, Albert Einstein College of Medicine, Bronx, NY 10461, USA, Department of Ophthalmology & Visual Sciences, Albert Einstein College of Medicine, Bronx, NY 10461, USA |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.ncbi.nlm.nih.gov/pmc/articles/5529005 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Challenges and progress in interpretation of non-coding genetic variants associated with human disease |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10222 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9991000294685364 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Genomics and Chromatin Dynamics |
| related_works | https://openalex.org/W3035076328, https://openalex.org/W1971424770, https://openalex.org/W2111842731, https://openalex.org/W2066467839, https://openalex.org/W2594299395, https://openalex.org/W2771541573, https://openalex.org/W2765794416, https://openalex.org/W2170506381, https://openalex.org/W2039666944, https://openalex.org/W1531630856 |
| cited_by_count | 70 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 4 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 22 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 11 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 4 |
| counts_by_year[4].year | 2021 |
| counts_by_year[4].cited_by_count | 6 |
| counts_by_year[5].year | 2020 |
| counts_by_year[5].cited_by_count | 13 |
| counts_by_year[6].year | 2019 |
| counts_by_year[6].cited_by_count | 8 |
| counts_by_year[7].year | 2018 |
| counts_by_year[7].cited_by_count | 1 |
| counts_by_year[8].year | 2017 |
| counts_by_year[8].cited_by_count | 1 |
| locations_count | 4 |
| best_oa_location.id | pmh:oai:pubmedcentral.nih.gov:5529005 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2764455111 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | PubMed Central |
| best_oa_location.source.host_organization | https://openalex.org/I1299303238 |
| best_oa_location.source.host_organization_name | National Institutes of Health |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I1299303238 |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | Text |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | Exp Biol Med (Maywood) |
| best_oa_location.landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/5529005 |
| primary_location.id | doi:10.1177/1535370217713750 |
| primary_location.is_oa | False |
| primary_location.source.id | https://openalex.org/S67514239 |
| primary_location.source.issn | 1535-3699, 1535-3702 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 1535-3699 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Experimental Biology and Medicine |
| primary_location.source.host_organization | https://openalex.org/P4310320017 |
| primary_location.source.host_organization_name | SAGE Publishing |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320017 |
| primary_location.source.host_organization_lineage_names | SAGE Publishing |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Experimental Biology and Medicine |
| primary_location.landing_page_url | https://doi.org/10.1177/1535370217713750 |
| publication_date | 2017-06-05 |
| publication_year | 2017 |
| referenced_works | https://openalex.org/W2121284504, https://openalex.org/W2116868464, https://openalex.org/W2118822739, https://openalex.org/W2147215182, https://openalex.org/W2142260323, https://openalex.org/W2156714600, https://openalex.org/W1636205509, https://openalex.org/W2555614465, https://openalex.org/W2209106767, https://openalex.org/W2065695399, https://openalex.org/W2097791641, https://openalex.org/W2259938310, https://openalex.org/W1844405070, https://openalex.org/W2159417227, https://openalex.org/W2067956561, https://openalex.org/W2076154138, https://openalex.org/W2014677321, https://openalex.org/W2152435464, https://openalex.org/W2002404684, https://openalex.org/W2014787267, https://openalex.org/W2015845481, https://openalex.org/W1996060670, https://openalex.org/W2168602111, https://openalex.org/W1981030303, https://openalex.org/W1531821806, https://openalex.org/W2149118429, https://openalex.org/W2091533196, https://openalex.org/W2030030642, https://openalex.org/W2140754284, https://openalex.org/W1983347147, https://openalex.org/W2084160423, https://openalex.org/W2063322714, https://openalex.org/W2145144631, https://openalex.org/W1977308882, https://openalex.org/W2118646732, https://openalex.org/W2021659821, https://openalex.org/W2128151873, https://openalex.org/W1971578941, https://openalex.org/W2004966039, https://openalex.org/W1969466735, https://openalex.org/W2082414039, https://openalex.org/W2060484997, https://openalex.org/W4213327946, https://openalex.org/W2117792884, https://openalex.org/W2056198580, https://openalex.org/W2077955816, https://openalex.org/W2125805216, https://openalex.org/W2016015848, https://openalex.org/W1900337607, https://openalex.org/W2018838463, https://openalex.org/W2077318478, https://openalex.org/W2195359644, https://openalex.org/W2141820415, https://openalex.org/W2951132213, https://openalex.org/W2018189081, https://openalex.org/W2160995259, https://openalex.org/W2136967659, https://openalex.org/W2023155906, https://openalex.org/W1993616000, https://openalex.org/W2138253007, https://openalex.org/W1964492509, https://openalex.org/W2602141149, https://openalex.org/W2023386301, https://openalex.org/W2077874719, https://openalex.org/W655820313, https://openalex.org/W2412481123, https://openalex.org/W2413629639, https://openalex.org/W2159664283, https://openalex.org/W2064815984, https://openalex.org/W2192824197, https://openalex.org/W2152505144, https://openalex.org/W2016991182, https://openalex.org/W2070708756, https://openalex.org/W2180291937, https://openalex.org/W1973260880, https://openalex.org/W2523099503, https://openalex.org/W1987320425, https://openalex.org/W1966781601, https://openalex.org/W2039176012, https://openalex.org/W2150904790, https://openalex.org/W2070509435, https://openalex.org/W1973062929, https://openalex.org/W1941076503, https://openalex.org/W2114866946, https://openalex.org/W1859117663, https://openalex.org/W2115464628, https://openalex.org/W2465545880, https://openalex.org/W2173155483, https://openalex.org/W1605145036, https://openalex.org/W2010789111, https://openalex.org/W1969208427, https://openalex.org/W2401262643, https://openalex.org/W2118246164, https://openalex.org/W2564368426, https://openalex.org/W2556214367, https://openalex.org/W2007988561, https://openalex.org/W2588079263 |
| referenced_works_count | 97 |
| abstract_inverted_index.a | 44, 148, 152 |
| abstract_inverted_index.By | 135 |
| abstract_inverted_index.To | 29 |
| abstract_inverted_index.We | 106 |
| abstract_inverted_index.be | 70 |
| abstract_inverted_index.in | 13, 65, 110 |
| abstract_inverted_index.is | 43 |
| abstract_inverted_index.no | 120 |
| abstract_inverted_index.of | 9, 17, 101, 112, 141 |
| abstract_inverted_index.to | 26, 47, 52, 72, 80, 94, 125, 155 |
| abstract_inverted_index.we | 60 |
| abstract_inverted_index.and | 35, 55, 64, 77, 98, 117, 139, 159 |
| abstract_inverted_index.but | 42 |
| abstract_inverted_index.can | 69 |
| abstract_inverted_index.far | 7 |
| abstract_inverted_index.for | 150 |
| abstract_inverted_index.map | 93 |
| abstract_inverted_index.one | 124 |
| abstract_inverted_index.our | 144 |
| abstract_inverted_index.the | 6, 14, 18, 49, 95, 114, 128, 137, 163 |
| abstract_inverted_index.GWAS | 133 |
| abstract_inverted_index.Here | 59 |
| abstract_inverted_index.Most | 86 |
| abstract_inverted_index.each | 142 |
| abstract_inverted_index.from | 88, 132 |
| abstract_inverted_index.gene | 22 |
| abstract_inverted_index.have | 3 |
| abstract_inverted_index.step | 46 |
| abstract_inverted_index.that | 5, 21, 68, 119 |
| abstract_inverted_index.used | 71 |
| abstract_inverted_index.genes | 39 |
| abstract_inverted_index.risk. | 28 |
| abstract_inverted_index.shown | 4 |
| abstract_inverted_index.their | 36, 82 |
| abstract_inverted_index.these | 102 |
| abstract_inverted_index.tools | 67 |
| abstract_inverted_index.truly | 31 |
| abstract_inverted_index.Impact | 84 |
| abstract_inverted_index.allows | 123 |
| abstract_inverted_index.argued | 118 |
| abstract_inverted_index.causal | 32, 74, 129, 164 |
| abstract_inverted_index.genome | 116 |
| abstract_inverted_index.recent | 108 |
| abstract_inverted_index.reside | 12 |
| abstract_inverted_index.review | 61, 145 |
| abstract_inverted_index.silico | 66 |
| abstract_inverted_index.single | 121 |
| abstract_inverted_index.taking | 151 |
| abstract_inverted_index.target | 38 |
| abstract_inverted_index.(GWASs) | 92 |
| abstract_inverted_index.changes | 24 |
| abstract_inverted_index.disease | 27 |
| abstract_inverted_index.genetic | 50 |
| abstract_inverted_index.genome, | 19, 97 |
| abstract_inverted_index.method, | 143 |
| abstract_inverted_index.regions | 16 |
| abstract_inverted_index.remains | 40 |
| abstract_inverted_index.signals | 87 |
| abstract_inverted_index.studies | 2, 91 |
| abstract_inverted_index.affected | 37 |
| abstract_inverted_index.approach | 122, 154 |
| abstract_inverted_index.clinical | 57 |
| abstract_inverted_index.critical | 45 |
| abstract_inverted_index.identify | 30, 73, 127 |
| abstract_inverted_index.majority | 8 |
| abstract_inverted_index.predict, | 157 |
| abstract_inverted_index.progress | 109 |
| abstract_inverted_index.provided | 147 |
| abstract_inverted_index.remained | 104 |
| abstract_inverted_index.results. | 134 |
| abstract_inverted_index.reviewed | 107 |
| abstract_inverted_index.studying | 113 |
| abstract_inverted_index.validate | 81, 162 |
| abstract_inverted_index.variants | 11, 34, 76, 131 |
| abstract_inverted_index.guideline | 149 |
| abstract_inverted_index.molecular | 53 |
| abstract_inverted_index.resources | 63 |
| abstract_inverted_index.statement | 85 |
| abstract_inverted_index.translate | 48 |
| abstract_inverted_index.variants. | 165 |
| abstract_inverted_index.accurately | 156 |
| abstract_inverted_index.advantages | 138 |
| abstract_inverted_index.contribute | 25 |
| abstract_inverted_index.eventually | 160 |
| abstract_inverted_index.functional | 99 |
| abstract_inverted_index.mechanisms | 54 |
| abstract_inverted_index.non-coding | 15, 33, 75, 96, 115 |
| abstract_inverted_index.regulatory | 23, 130 |
| abstract_inverted_index.strategies | 79 |
| abstract_inverted_index.suggesting | 20 |
| abstract_inverted_index.ultimately | 56 |
| abstract_inverted_index.Genome-wide | 0 |
| abstract_inverted_index.association | 1, 90 |
| abstract_inverted_index.challenging | 41 |
| abstract_inverted_index.effectively | 126 |
| abstract_inverted_index.genome-wide | 89 |
| abstract_inverted_index.limitations | 140 |
| abstract_inverted_index.potentially | 146 |
| abstract_inverted_index.prioritize, | 158 |
| abstract_inverted_index.associations | 51, 103 |
| abstract_inverted_index.challenging. | 105 |
| abstract_inverted_index.experimental | 78 |
| abstract_inverted_index.illustrating | 136 |
| abstract_inverted_index.applications. | 58 |
| abstract_inverted_index.combinatorial | 153 |
| abstract_inverted_index.methodologies | 111 |
| abstract_inverted_index.experimentally | 161 |
| abstract_inverted_index.interpretation | 100 |
| abstract_inverted_index.functionalities. | 83 |
| abstract_inverted_index.disease-associated | 10 |
| abstract_inverted_index.genomic/epigenomic | 62 |
| cited_by_percentile_year.max | 100 |
| cited_by_percentile_year.min | 90 |
| corresponding_author_ids | https://openalex.org/A5080863597 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| corresponding_institution_ids | https://openalex.org/I129975664 |
| citation_normalized_percentile.value | 0.89962594 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |