Choreography of budding yeast chromosomes during the cell cycle Article Swipe
 YOU?
·
· 2016
· Open Access
·
· DOI: https://doi.org/10.1101/096826
YOU?
·
· 2016
· Open Access
·
· DOI: https://doi.org/10.1101/096826
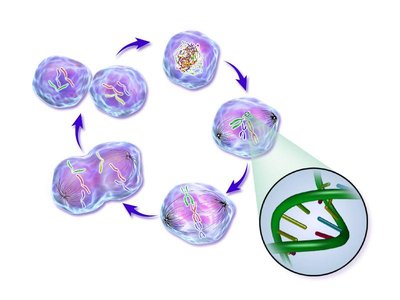
To ensure the proper transmission of the genetic information, DNA molecules must be faithfully duplicated and segregated. These processes involve dynamic modifications of chromosomes internal structure to promote their individualization, as well as their global repositioning into daughter cells (Guacci et al., 1994; Kleckner et al., 2014; Mizuguchi et al., 2014). In eukaryotes, these events are regulated by conserved architectural proteins, such as structural maintenance of chromosomes (SMC i.e. cohesin and condensin) complexes (Aragon et al., 2013a; Uhlmann, 2016). Although the roles of these factors have been actively investigated, the genome-wide chromosomal architecture and dynamics both at small and large-scales during cell division remains elusive. Here we report a comprehensive Hi-C (Dekker et al., 2002; Lieberman-Aiden et al., 2009) analysis of the dynamic changes of chromosomes structure over the Saccharomyces cerevisiae cell cycle. We uncover specific SMC-dependent structural transitions between the different phases of the mitotic cycle. During replication, cohesion establishment promotes the increase of long-range intra-chromosomal contacts. This process correlates with the individualization of chromosomes, which culminates at metaphase. Mitotic chromosomes are then abruptly reorganized in anaphase by the mechanical forces exerted by the mitotic spindle on the centromere cluster. The formation of a condensin-dependent loop, that bridges centromere cluster with the cenproximal flanking region of the rDNA, suggests that these forces may directly facilitate nucleolus segregation. This work provides a comprehensive overview of chromosome dynamics during the cell cycle of a unicellular eukaryote that recapitulates and unveils new features of highly conserved stages of the cell division.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/096826
- https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdf
- OA Status
- green
- Cited By
- 7
- References
- 36
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2561883008
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2561883008Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/096826Digital Object Identifier
- Title
-
Choreography of budding yeast chromosomes during the cell cycleWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2016Year of publication
- Publication date
-
2016-12-30Full publication date if available
- Authors
-
Luciana Lazar‐Stefanita, Vittore F. Scolari, Guillaume Mercy, Agnès Thierry, Héloïse Muller, Julien Mozziconacci, Romain KoszulList of authors in order
- Landing page
-
https://doi.org/10.1101/096826Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdfDirect OA link when available
- Concepts
-
Cohesin, Biology, Centromere, Chromosome segregation, Mitosis, Condensin, Anaphase, Genetics, Cell cycle, Chromatin, Cell biology, Metaphase, Cell division, Genome, Chromosome, DNA, Cell, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
7Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2022: 1, 2019: 2, 2018: 1, 2017: 2Per-year citation counts (last 5 years)
- References (count)
-
36Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2561883008 |
|---|---|
| doi | https://doi.org/10.1101/096826 |
| ids.doi | https://doi.org/10.1101/096826 |
| ids.mag | 2561883008 |
| ids.openalex | https://openalex.org/W2561883008 |
| fwci | 0.86663083 |
| type | preprint |
| title | Choreography of budding yeast chromosomes during the cell cycle |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10222 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Genomics and Chromatin Dynamics |
| topics[1].id | https://openalex.org/T10434 |
| topics[1].field.id | https://openalex.org/fields/11 |
| topics[1].field.display_name | Agricultural and Biological Sciences |
| topics[1].score | 0.9991000294685364 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1110 |
| topics[1].subfield.display_name | Plant Science |
| topics[1].display_name | Chromosomal and Genetic Variations |
| topics[2].id | https://openalex.org/T10492 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.998199999332428 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1307 |
| topics[2].subfield.display_name | Cell Biology |
| topics[2].display_name | Microtubule and mitosis dynamics |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C115821613 |
| concepts[0].level | 4 |
| concepts[0].score | 0.85460364818573 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21103063 |
| concepts[0].display_name | Cohesin |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.7204842567443848 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C63491729 |
| concepts[2].level | 4 |
| concepts[2].score | 0.6880483627319336 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q14762596 |
| concepts[2].display_name | Centromere |
| concepts[3].id | https://openalex.org/C2778354886 |
| concepts[3].level | 4 |
| concepts[3].score | 0.6317338347434998 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q5113927 |
| concepts[3].display_name | Chromosome segregation |
| concepts[4].id | https://openalex.org/C93304396 |
| concepts[4].level | 2 |
| concepts[4].score | 0.6183218359947205 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q14763008 |
| concepts[4].display_name | Mitosis |
| concepts[5].id | https://openalex.org/C155790170 |
| concepts[5].level | 5 |
| concepts[5].score | 0.5952274203300476 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q21105628 |
| concepts[5].display_name | Condensin |
| concepts[6].id | https://openalex.org/C53420372 |
| concepts[6].level | 4 |
| concepts[6].score | 0.5592557191848755 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q22299157 |
| concepts[6].display_name | Anaphase |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.5091366171836853 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C29537977 |
| concepts[8].level | 3 |
| concepts[8].score | 0.4953412115573883 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q188941 |
| concepts[8].display_name | Cell cycle |
| concepts[9].id | https://openalex.org/C83640560 |
| concepts[9].level | 3 |
| concepts[9].score | 0.48836657404899597 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q180951 |
| concepts[9].display_name | Chromatin |
| concepts[10].id | https://openalex.org/C95444343 |
| concepts[10].level | 1 |
| concepts[10].score | 0.47756439447402954 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[10].display_name | Cell biology |
| concepts[11].id | https://openalex.org/C70278784 |
| concepts[11].level | 4 |
| concepts[11].score | 0.44545862078666687 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q574427 |
| concepts[11].display_name | Metaphase |
| concepts[12].id | https://openalex.org/C85813293 |
| concepts[12].level | 3 |
| concepts[12].score | 0.43803125619888306 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q188909 |
| concepts[12].display_name | Cell division |
| concepts[13].id | https://openalex.org/C141231307 |
| concepts[13].level | 3 |
| concepts[13].score | 0.4176047444343567 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[13].display_name | Genome |
| concepts[14].id | https://openalex.org/C30481170 |
| concepts[14].level | 3 |
| concepts[14].score | 0.40956783294677734 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q37748 |
| concepts[14].display_name | Chromosome |
| concepts[15].id | https://openalex.org/C552990157 |
| concepts[15].level | 2 |
| concepts[15].score | 0.2374681532382965 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[15].display_name | DNA |
| concepts[16].id | https://openalex.org/C1491633281 |
| concepts[16].level | 2 |
| concepts[16].score | 0.1828039288520813 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7868 |
| concepts[16].display_name | Cell |
| concepts[17].id | https://openalex.org/C104317684 |
| concepts[17].level | 2 |
| concepts[17].score | 0.09634515643119812 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[17].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/cohesin |
| keywords[0].score | 0.85460364818573 |
| keywords[0].display_name | Cohesin |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.7204842567443848 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/centromere |
| keywords[2].score | 0.6880483627319336 |
| keywords[2].display_name | Centromere |
| keywords[3].id | https://openalex.org/keywords/chromosome-segregation |
| keywords[3].score | 0.6317338347434998 |
| keywords[3].display_name | Chromosome segregation |
| keywords[4].id | https://openalex.org/keywords/mitosis |
| keywords[4].score | 0.6183218359947205 |
| keywords[4].display_name | Mitosis |
| keywords[5].id | https://openalex.org/keywords/condensin |
| keywords[5].score | 0.5952274203300476 |
| keywords[5].display_name | Condensin |
| keywords[6].id | https://openalex.org/keywords/anaphase |
| keywords[6].score | 0.5592557191848755 |
| keywords[6].display_name | Anaphase |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.5091366171836853 |
| keywords[7].display_name | Genetics |
| keywords[8].id | https://openalex.org/keywords/cell-cycle |
| keywords[8].score | 0.4953412115573883 |
| keywords[8].display_name | Cell cycle |
| keywords[9].id | https://openalex.org/keywords/chromatin |
| keywords[9].score | 0.48836657404899597 |
| keywords[9].display_name | Chromatin |
| keywords[10].id | https://openalex.org/keywords/cell-biology |
| keywords[10].score | 0.47756439447402954 |
| keywords[10].display_name | Cell biology |
| keywords[11].id | https://openalex.org/keywords/metaphase |
| keywords[11].score | 0.44545862078666687 |
| keywords[11].display_name | Metaphase |
| keywords[12].id | https://openalex.org/keywords/cell-division |
| keywords[12].score | 0.43803125619888306 |
| keywords[12].display_name | Cell division |
| keywords[13].id | https://openalex.org/keywords/genome |
| keywords[13].score | 0.4176047444343567 |
| keywords[13].display_name | Genome |
| keywords[14].id | https://openalex.org/keywords/chromosome |
| keywords[14].score | 0.40956783294677734 |
| keywords[14].display_name | Chromosome |
| keywords[15].id | https://openalex.org/keywords/dna |
| keywords[15].score | 0.2374681532382965 |
| keywords[15].display_name | DNA |
| keywords[16].id | https://openalex.org/keywords/cell |
| keywords[16].score | 0.1828039288520813 |
| keywords[16].display_name | Cell |
| keywords[17].id | https://openalex.org/keywords/gene |
| keywords[17].score | 0.09634515643119812 |
| keywords[17].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1101/096826 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/096826 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5026571760 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-2388-9757 |
| authorships[0].author.display_name | Luciana Lazar‐Stefanita |
| authorships[0].countries | FR |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I157536573 |
| authorships[0].affiliations[0].raw_affiliation_string | Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I1294671590 |
| authorships[0].affiliations[1].raw_affiliation_string | CNRS, UMR 355, 75015 Paris, France |
| authorships[0].affiliations[2].institution_ids | https://openalex.org/I157536573 |
| authorships[0].affiliations[2].raw_affiliation_string | Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[0].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[0].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[0].institutions[0].type | government |
| authorships[0].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[0].institutions[0].country_code | FR |
| authorships[0].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[0].institutions[1].id | https://openalex.org/I157536573 |
| authorships[0].institutions[1].ror | https://ror.org/0495fxg12 |
| authorships[0].institutions[1].type | nonprofit |
| authorships[0].institutions[1].lineage | https://openalex.org/I157536573 |
| authorships[0].institutions[1].country_code | FR |
| authorships[0].institutions[1].display_name | Institut Pasteur |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Luciana Lazar-Stefanita |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | CNRS, UMR 355, 75015 Paris, France, Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France, Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[1].author.id | https://openalex.org/A5058832006 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3490-0579 |
| authorships[1].author.display_name | Vittore F. Scolari |
| authorships[1].countries | FR |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I157536573 |
| authorships[1].affiliations[0].raw_affiliation_string | Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I1294671590 |
| authorships[1].affiliations[1].raw_affiliation_string | CNRS, UMR 355, 75015 Paris, France |
| authorships[1].affiliations[2].institution_ids | https://openalex.org/I157536573 |
| authorships[1].affiliations[2].raw_affiliation_string | Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[1].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[1].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[1].institutions[0].type | government |
| authorships[1].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[1].institutions[0].country_code | FR |
| authorships[1].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[1].institutions[1].id | https://openalex.org/I157536573 |
| authorships[1].institutions[1].ror | https://ror.org/0495fxg12 |
| authorships[1].institutions[1].type | nonprofit |
| authorships[1].institutions[1].lineage | https://openalex.org/I157536573 |
| authorships[1].institutions[1].country_code | FR |
| authorships[1].institutions[1].display_name | Institut Pasteur |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Vittore F. Scolari |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | CNRS, UMR 355, 75015 Paris, France, Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France, Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[2].author.id | https://openalex.org/A5037970725 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-6274-5927 |
| authorships[2].author.display_name | Guillaume Mercy |
| authorships[2].countries | FR |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I157536573 |
| authorships[2].affiliations[0].raw_affiliation_string | Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I1294671590 |
| authorships[2].affiliations[1].raw_affiliation_string | CNRS, UMR 355, 75015 Paris, France |
| authorships[2].affiliations[2].institution_ids | https://openalex.org/I157536573 |
| authorships[2].affiliations[2].raw_affiliation_string | Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[2].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[2].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[2].institutions[0].type | government |
| authorships[2].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[2].institutions[0].country_code | FR |
| authorships[2].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[2].institutions[1].id | https://openalex.org/I157536573 |
| authorships[2].institutions[1].ror | https://ror.org/0495fxg12 |
| authorships[2].institutions[1].type | nonprofit |
| authorships[2].institutions[1].lineage | https://openalex.org/I157536573 |
| authorships[2].institutions[1].country_code | FR |
| authorships[2].institutions[1].display_name | Institut Pasteur |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Guillaume Mercy |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | CNRS, UMR 355, 75015 Paris, France, Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France, Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[3].author.id | https://openalex.org/A5107849334 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Agnès Thierry |
| authorships[3].countries | FR |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I157536573 |
| authorships[3].affiliations[0].raw_affiliation_string | Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I1294671590 |
| authorships[3].affiliations[1].raw_affiliation_string | CNRS, UMR 355, 75015 Paris, France |
| authorships[3].affiliations[2].institution_ids | https://openalex.org/I157536573 |
| authorships[3].affiliations[2].raw_affiliation_string | Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[3].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[3].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[3].institutions[0].type | government |
| authorships[3].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[3].institutions[0].country_code | FR |
| authorships[3].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[3].institutions[1].id | https://openalex.org/I157536573 |
| authorships[3].institutions[1].ror | https://ror.org/0495fxg12 |
| authorships[3].institutions[1].type | nonprofit |
| authorships[3].institutions[1].lineage | https://openalex.org/I157536573 |
| authorships[3].institutions[1].country_code | FR |
| authorships[3].institutions[1].display_name | Institut Pasteur |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Agnès Thierry |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | CNRS, UMR 355, 75015 Paris, France, Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France, Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[4].author.id | https://openalex.org/A5102990753 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-0853-822X |
| authorships[4].author.display_name | Héloïse Muller |
| authorships[4].countries | FR |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I1294671590 |
| authorships[4].affiliations[0].raw_affiliation_string | CNRS, UMR 355, 75015 Paris, France |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I157536573 |
| authorships[4].affiliations[1].raw_affiliation_string | Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[4].affiliations[2].institution_ids | https://openalex.org/I157536573 |
| authorships[4].affiliations[2].raw_affiliation_string | Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France |
| authorships[4].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[4].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[4].institutions[0].type | government |
| authorships[4].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[4].institutions[0].country_code | FR |
| authorships[4].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[4].institutions[1].id | https://openalex.org/I157536573 |
| authorships[4].institutions[1].ror | https://ror.org/0495fxg12 |
| authorships[4].institutions[1].type | nonprofit |
| authorships[4].institutions[1].lineage | https://openalex.org/I157536573 |
| authorships[4].institutions[1].country_code | FR |
| authorships[4].institutions[1].display_name | Institut Pasteur |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Héloise Muller |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | CNRS, UMR 355, 75015 Paris, France, Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France, Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[5].author.id | https://openalex.org/A5011027517 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-5652-0302 |
| authorships[5].author.display_name | Julien Mozziconacci |
| authorships[5].countries | FR |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I1294671590 |
| authorships[5].affiliations[0].raw_affiliation_string | CNRS, UMR 700, 75005 Paris, France |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I39804081, https://openalex.org/I4210140797 |
| authorships[5].affiliations[1].raw_affiliation_string | Sorbonne Universités, Theoretical Physics for condensed matter lab, UPMC Université Paris 06, 700 Paris, France |
| authorships[5].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[5].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[5].institutions[0].type | government |
| authorships[5].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[5].institutions[0].country_code | FR |
| authorships[5].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[5].institutions[1].id | https://openalex.org/I4210140797 |
| authorships[5].institutions[1].ror | https://ror.org/04zaaa143 |
| authorships[5].institutions[1].type | facility |
| authorships[5].institutions[1].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I39804081, https://openalex.org/I4210098836, https://openalex.org/I4210140797 |
| authorships[5].institutions[1].country_code | FR |
| authorships[5].institutions[1].display_name | Laboratoire de Physique Théorique de la Matière Condensée |
| authorships[5].institutions[2].id | https://openalex.org/I39804081 |
| authorships[5].institutions[2].ror | https://ror.org/02en5vm52 |
| authorships[5].institutions[2].type | education |
| authorships[5].institutions[2].lineage | https://openalex.org/I39804081 |
| authorships[5].institutions[2].country_code | FR |
| authorships[5].institutions[2].display_name | Sorbonne Université |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Julien Mozziconacci |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | CNRS, UMR 700, 75005 Paris, France, Sorbonne Universités, Theoretical Physics for condensed matter lab, UPMC Université Paris 06, 700 Paris, France |
| authorships[6].author.id | https://openalex.org/A5045736877 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-3086-1173 |
| authorships[6].author.display_name | Romain Koszul |
| authorships[6].countries | FR |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1294671590 |
| authorships[6].affiliations[0].raw_affiliation_string | CNRS, UMR 355, 75015 Paris, France |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I157536573 |
| authorships[6].affiliations[1].raw_affiliation_string | Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| authorships[6].affiliations[2].institution_ids | https://openalex.org/I157536573 |
| authorships[6].affiliations[2].raw_affiliation_string | Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France |
| authorships[6].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[6].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[6].institutions[0].type | government |
| authorships[6].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[6].institutions[0].country_code | FR |
| authorships[6].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[6].institutions[1].id | https://openalex.org/I157536573 |
| authorships[6].institutions[1].ror | https://ror.org/0495fxg12 |
| authorships[6].institutions[1].type | nonprofit |
| authorships[6].institutions[1].lineage | https://openalex.org/I157536573 |
| authorships[6].institutions[1].country_code | FR |
| authorships[6].institutions[1].display_name | Institut Pasteur |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Romain Koszul |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | CNRS, UMR 355, 75015 Paris, France, Institut Pasteur, Center of Bioinformatics, Biostatistics and Integrative Biology (CBI),Paris, F-75015, France, Institut Pasteur, Department Genomes and Genetics, Groupe Régulation Spatiale des Génomes, 7505 Paris, France |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Choreography of budding yeast chromosomes during the cell cycle |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10222 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Genomics and Chromatin Dynamics |
| related_works | https://openalex.org/W1964977825, https://openalex.org/W2784825908, https://openalex.org/W2793314979, https://openalex.org/W2125659069, https://openalex.org/W2117256838, https://openalex.org/W1975394594, https://openalex.org/W4238425513, https://openalex.org/W2745958356, https://openalex.org/W2080113789, https://openalex.org/W3119633924 |
| cited_by_count | 7 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2022 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2019 |
| counts_by_year[2].cited_by_count | 2 |
| counts_by_year[3].year | 2018 |
| counts_by_year[3].cited_by_count | 1 |
| counts_by_year[4].year | 2017 |
| counts_by_year[4].cited_by_count | 2 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/096826 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/096826 |
| primary_location.id | doi:10.1101/096826 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2016/12/30/096826.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/096826 |
| publication_date | 2016-12-30 |
| publication_year | 2016 |
| referenced_works | https://openalex.org/W2072257534, https://openalex.org/W2152605077, https://openalex.org/W2133188555, https://openalex.org/W2066833380, https://openalex.org/W2150455380, https://openalex.org/W2137507610, https://openalex.org/W2051893661, https://openalex.org/W2073026107, https://openalex.org/W2069721588, https://openalex.org/W2142157138, https://openalex.org/W2129835359, https://openalex.org/W2136202460, https://openalex.org/W2159137674, https://openalex.org/W2137856365, https://openalex.org/W2051018998, https://openalex.org/W2029667189, https://openalex.org/W2133112673, https://openalex.org/W1994919128, https://openalex.org/W1984637914, https://openalex.org/W1994536648, https://openalex.org/W2090037139, https://openalex.org/W1515246058, https://openalex.org/W2106864638, https://openalex.org/W2099357999, https://openalex.org/W2416606897, https://openalex.org/W2073504534, https://openalex.org/W1579608990, https://openalex.org/W2008486131, https://openalex.org/W2168463084, https://openalex.org/W2102784353, https://openalex.org/W2126026810, https://openalex.org/W2093837256, https://openalex.org/W2341542582, https://openalex.org/W1638194466, https://openalex.org/W2006008512, https://openalex.org/W2127926172 |
| referenced_works_count | 36 |
| abstract_inverted_index.a | 109, 195, 222, 233 |
| abstract_inverted_index.In | 52 |
| abstract_inverted_index.To | 1 |
| abstract_inverted_index.We | 134 |
| abstract_inverted_index.as | 31, 33, 63 |
| abstract_inverted_index.at | 97, 169 |
| abstract_inverted_index.be | 13 |
| abstract_inverted_index.by | 58, 179, 184 |
| abstract_inverted_index.et | 41, 45, 49, 75, 113, 117 |
| abstract_inverted_index.in | 177 |
| abstract_inverted_index.of | 6, 23, 66, 83, 121, 125, 144, 155, 165, 194, 207, 225, 232, 242, 246 |
| abstract_inverted_index.on | 188 |
| abstract_inverted_index.to | 27 |
| abstract_inverted_index.we | 107 |
| abstract_inverted_index.DNA | 10 |
| abstract_inverted_index.The | 192 |
| abstract_inverted_index.and | 16, 71, 94, 99, 238 |
| abstract_inverted_index.are | 56, 173 |
| abstract_inverted_index.may | 214 |
| abstract_inverted_index.new | 240 |
| abstract_inverted_index.the | 3, 7, 81, 90, 122, 129, 141, 145, 153, 163, 180, 185, 189, 203, 208, 229, 247 |
| abstract_inverted_index.(SMC | 68 |
| abstract_inverted_index.Here | 106 |
| abstract_inverted_index.Hi-C | 111 |
| abstract_inverted_index.This | 159, 219 |
| abstract_inverted_index.al., | 42, 46, 50, 76, 114, 118 |
| abstract_inverted_index.been | 87 |
| abstract_inverted_index.both | 96 |
| abstract_inverted_index.cell | 102, 132, 230, 248 |
| abstract_inverted_index.have | 86 |
| abstract_inverted_index.i.e. | 69 |
| abstract_inverted_index.into | 37 |
| abstract_inverted_index.must | 12 |
| abstract_inverted_index.over | 128 |
| abstract_inverted_index.such | 62 |
| abstract_inverted_index.that | 198, 211, 236 |
| abstract_inverted_index.then | 174 |
| abstract_inverted_index.well | 32 |
| abstract_inverted_index.with | 162, 202 |
| abstract_inverted_index.work | 220 |
| abstract_inverted_index.1994; | 43 |
| abstract_inverted_index.2002; | 115 |
| abstract_inverted_index.2009) | 119 |
| abstract_inverted_index.2014; | 47 |
| abstract_inverted_index.These | 18 |
| abstract_inverted_index.cells | 39 |
| abstract_inverted_index.cycle | 231 |
| abstract_inverted_index.loop, | 197 |
| abstract_inverted_index.rDNA, | 209 |
| abstract_inverted_index.roles | 82 |
| abstract_inverted_index.small | 98 |
| abstract_inverted_index.their | 29, 34 |
| abstract_inverted_index.these | 54, 84, 212 |
| abstract_inverted_index.which | 167 |
| abstract_inverted_index.2013a; | 77 |
| abstract_inverted_index.2014). | 51 |
| abstract_inverted_index.2016). | 79 |
| abstract_inverted_index.During | 148 |
| abstract_inverted_index.cycle. | 133, 147 |
| abstract_inverted_index.during | 101, 228 |
| abstract_inverted_index.ensure | 2 |
| abstract_inverted_index.events | 55 |
| abstract_inverted_index.forces | 182, 213 |
| abstract_inverted_index.global | 35 |
| abstract_inverted_index.highly | 243 |
| abstract_inverted_index.phases | 143 |
| abstract_inverted_index.proper | 4 |
| abstract_inverted_index.region | 206 |
| abstract_inverted_index.report | 108 |
| abstract_inverted_index.stages | 245 |
| abstract_inverted_index.(Aragon | 74 |
| abstract_inverted_index.(Dekker | 112 |
| abstract_inverted_index.(Guacci | 40 |
| abstract_inverted_index.Mitotic | 171 |
| abstract_inverted_index.between | 140 |
| abstract_inverted_index.bridges | 199 |
| abstract_inverted_index.changes | 124 |
| abstract_inverted_index.cluster | 201 |
| abstract_inverted_index.cohesin | 70 |
| abstract_inverted_index.dynamic | 21, 123 |
| abstract_inverted_index.exerted | 183 |
| abstract_inverted_index.factors | 85 |
| abstract_inverted_index.genetic | 8 |
| abstract_inverted_index.involve | 20 |
| abstract_inverted_index.mitotic | 146, 186 |
| abstract_inverted_index.process | 160 |
| abstract_inverted_index.promote | 28 |
| abstract_inverted_index.remains | 104 |
| abstract_inverted_index.spindle | 187 |
| abstract_inverted_index.uncover | 135 |
| abstract_inverted_index.unveils | 239 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Although | 80 |
| abstract_inverted_index.Kleckner | 44 |
| abstract_inverted_index.Uhlmann, | 78 |
| abstract_inverted_index.abruptly | 175 |
| abstract_inverted_index.actively | 88 |
| abstract_inverted_index.analysis | 120 |
| abstract_inverted_index.anaphase | 178 |
| abstract_inverted_index.cluster. | 191 |
| abstract_inverted_index.cohesion | 150 |
| abstract_inverted_index.daughter | 38 |
| abstract_inverted_index.directly | 215 |
| abstract_inverted_index.division | 103 |
| abstract_inverted_index.dynamics | 95, 227 |
| abstract_inverted_index.elusive. | 105 |
| abstract_inverted_index.features | 241 |
| abstract_inverted_index.flanking | 205 |
| abstract_inverted_index.increase | 154 |
| abstract_inverted_index.internal | 25 |
| abstract_inverted_index.overview | 224 |
| abstract_inverted_index.promotes | 152 |
| abstract_inverted_index.provides | 221 |
| abstract_inverted_index.specific | 136 |
| abstract_inverted_index.suggests | 210 |
| abstract_inverted_index.Mizuguchi | 48 |
| abstract_inverted_index.complexes | 73 |
| abstract_inverted_index.conserved | 59, 244 |
| abstract_inverted_index.contacts. | 158 |
| abstract_inverted_index.different | 142 |
| abstract_inverted_index.division. | 249 |
| abstract_inverted_index.eukaryote | 235 |
| abstract_inverted_index.formation | 193 |
| abstract_inverted_index.molecules | 11 |
| abstract_inverted_index.nucleolus | 217 |
| abstract_inverted_index.processes | 19 |
| abstract_inverted_index.proteins, | 61 |
| abstract_inverted_index.regulated | 57 |
| abstract_inverted_index.structure | 26, 127 |
| abstract_inverted_index.centromere | 190, 200 |
| abstract_inverted_index.cerevisiae | 131 |
| abstract_inverted_index.chromosome | 226 |
| abstract_inverted_index.condensin) | 72 |
| abstract_inverted_index.correlates | 161 |
| abstract_inverted_index.culminates | 168 |
| abstract_inverted_index.duplicated | 15 |
| abstract_inverted_index.facilitate | 216 |
| abstract_inverted_index.faithfully | 14 |
| abstract_inverted_index.long-range | 156 |
| abstract_inverted_index.mechanical | 181 |
| abstract_inverted_index.metaphase. | 170 |
| abstract_inverted_index.structural | 64, 138 |
| abstract_inverted_index.cenproximal | 204 |
| abstract_inverted_index.chromosomal | 92 |
| abstract_inverted_index.chromosomes | 24, 67, 126, 172 |
| abstract_inverted_index.eukaryotes, | 53 |
| abstract_inverted_index.genome-wide | 91 |
| abstract_inverted_index.maintenance | 65 |
| abstract_inverted_index.reorganized | 176 |
| abstract_inverted_index.segregated. | 17 |
| abstract_inverted_index.transitions | 139 |
| abstract_inverted_index.unicellular | 234 |
| abstract_inverted_index.architecture | 93 |
| abstract_inverted_index.chromosomes, | 166 |
| abstract_inverted_index.information, | 9 |
| abstract_inverted_index.large-scales | 100 |
| abstract_inverted_index.replication, | 149 |
| abstract_inverted_index.segregation. | 218 |
| abstract_inverted_index.transmission | 5 |
| abstract_inverted_index.SMC-dependent | 137 |
| abstract_inverted_index.Saccharomyces | 130 |
| abstract_inverted_index.architectural | 60 |
| abstract_inverted_index.comprehensive | 110, 223 |
| abstract_inverted_index.establishment | 151 |
| abstract_inverted_index.investigated, | 89 |
| abstract_inverted_index.modifications | 22 |
| abstract_inverted_index.recapitulates | 237 |
| abstract_inverted_index.repositioning | 36 |
| abstract_inverted_index.Lieberman-Aiden | 116 |
| abstract_inverted_index.individualization | 164 |
| abstract_inverted_index.intra-chromosomal | 157 |
| abstract_inverted_index.individualization, | 30 |
| abstract_inverted_index.condensin-dependent | 196 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 89 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 7 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.47999998927116394 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.78939755 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |