Classification of All Blood Cell Images using ML and DL Models Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.48550/arxiv.2308.06300
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.48550/arxiv.2308.06300
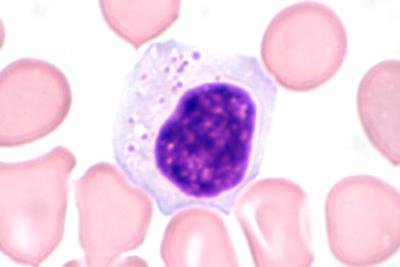
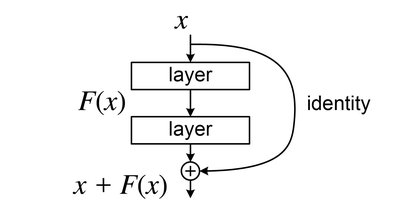
Human blood primarily comprises plasma, red blood cells, white blood cells, and platelets. It plays a vital role in transporting nutrients to different organs, where it stores essential health-related data about the human body. Blood cells are utilized to defend the body against diverse infections, including fungi, viruses, and bacteria. Hence, blood analysis can help physicians assess an individual's physiological condition. Blood cells have been sub-classified into eight groups: Neutrophils, eosinophils, basophils, lymphocytes, monocytes, immature granulocytes (promyelocytes, myelocytes, and metamyelocytes), erythroblasts, and platelets or thrombocytes on the basis of their nucleus, shape, and cytoplasm. Traditionally, pathologists and hematologists in laboratories have examined these blood cells using a microscope before manually classifying them. The manual approach is slower and more prone to human error. Therefore, it is essential to automate this process. In our paper, transfer learning with CNN pre-trained models. VGG16, VGG19, ResNet-50, ResNet-101, ResNet-152, InceptionV3, MobileNetV2, and DenseNet-20 applied to the PBC dataset's normal DIB. The overall accuracy achieved with these models lies between 91.375 and 94.72%. Hence, inspired by these pre-trained architectures, a model has been proposed to automatically classify the ten types of blood cells with increased accuracy. A novel CNN-based framework has been presented to improve accuracy. The proposed CNN model has been tested on the PBC dataset normal DIB. The outcomes of the experiments demonstrate that our CNN-based framework designed for blood cell classification attains an accuracy of 99.91% on the PBC dataset. Our proposed convolutional neural network model performs competitively when compared to earlier results reported in the literature.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- http://arxiv.org/abs/2308.06300
- https://arxiv.org/pdf/2308.06300
- OA Status
- green
- Cited By
- 2
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4385848370
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4385848370Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.48550/arxiv.2308.06300Digital Object Identifier
- Title
-
Classification of All Blood Cell Images using ML and DL ModelsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-08-11Full publication date if available
- Authors
-
Rabia Asghar, Sanjay Kumar, Abeera MahfoozList of authors in order
- Landing page
-
https://arxiv.org/abs/2308.06300Publisher landing page
- PDF URL
-
https://arxiv.org/pdf/2308.06300Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://arxiv.org/pdf/2308.06300Direct OA link when available
- Concepts
-
Convolutional neural network, Transfer of learning, Platelet, Blood cell, Artificial intelligence, White blood cell, Deep learning, Human blood, Residual neural network, Computer science, Pattern recognition (psychology), Immunology, Medicine, PhysiologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
2Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 2Per-year citation counts (last 5 years)
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4385848370 |
|---|---|
| doi | https://doi.org/10.48550/arxiv.2308.06300 |
| ids.doi | https://doi.org/10.48550/arxiv.2308.06300 |
| ids.openalex | https://openalex.org/W4385848370 |
| fwci | 0.36393668 |
| type | preprint |
| title | Classification of All Blood Cell Images using ML and DL Models |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T12874 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1707 |
| topics[0].subfield.display_name | Computer Vision and Pattern Recognition |
| topics[0].display_name | Digital Imaging for Blood Diseases |
| topics[1].id | https://openalex.org/T11775 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9527999758720398 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2741 |
| topics[1].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[1].display_name | COVID-19 diagnosis using AI |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C81363708 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8280615210533142 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q17084460 |
| concepts[0].display_name | Convolutional neural network |
| concepts[1].id | https://openalex.org/C150899416 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5972819328308105 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1820378 |
| concepts[1].display_name | Transfer of learning |
| concepts[2].id | https://openalex.org/C89560881 |
| concepts[2].level | 2 |
| concepts[2].score | 0.581804633140564 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q101026 |
| concepts[2].display_name | Platelet |
| concepts[3].id | https://openalex.org/C2779979121 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5378889441490173 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q211709 |
| concepts[3].display_name | Blood cell |
| concepts[4].id | https://openalex.org/C154945302 |
| concepts[4].level | 1 |
| concepts[4].score | 0.532296895980835 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[4].display_name | Artificial intelligence |
| concepts[5].id | https://openalex.org/C2778488018 |
| concepts[5].level | 2 |
| concepts[5].score | 0.514819324016571 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q42395 |
| concepts[5].display_name | White blood cell |
| concepts[6].id | https://openalex.org/C108583219 |
| concepts[6].level | 2 |
| concepts[6].score | 0.5147202610969543 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q197536 |
| concepts[6].display_name | Deep learning |
| concepts[7].id | https://openalex.org/C3020437955 |
| concepts[7].level | 2 |
| concepts[7].score | 0.47320979833602905 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7873 |
| concepts[7].display_name | Human blood |
| concepts[8].id | https://openalex.org/C2944601119 |
| concepts[8].level | 3 |
| concepts[8].score | 0.4637741446495056 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q43744058 |
| concepts[8].display_name | Residual neural network |
| concepts[9].id | https://openalex.org/C41008148 |
| concepts[9].level | 0 |
| concepts[9].score | 0.460194855928421 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[9].display_name | Computer science |
| concepts[10].id | https://openalex.org/C153180895 |
| concepts[10].level | 2 |
| concepts[10].score | 0.4227150082588196 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7148389 |
| concepts[10].display_name | Pattern recognition (psychology) |
| concepts[11].id | https://openalex.org/C203014093 |
| concepts[11].level | 1 |
| concepts[11].score | 0.3050536513328552 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[11].display_name | Immunology |
| concepts[12].id | https://openalex.org/C71924100 |
| concepts[12].level | 0 |
| concepts[12].score | 0.278763085603714 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[12].display_name | Medicine |
| concepts[13].id | https://openalex.org/C42407357 |
| concepts[13].level | 1 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q521 |
| concepts[13].display_name | Physiology |
| keywords[0].id | https://openalex.org/keywords/convolutional-neural-network |
| keywords[0].score | 0.8280615210533142 |
| keywords[0].display_name | Convolutional neural network |
| keywords[1].id | https://openalex.org/keywords/transfer-of-learning |
| keywords[1].score | 0.5972819328308105 |
| keywords[1].display_name | Transfer of learning |
| keywords[2].id | https://openalex.org/keywords/platelet |
| keywords[2].score | 0.581804633140564 |
| keywords[2].display_name | Platelet |
| keywords[3].id | https://openalex.org/keywords/blood-cell |
| keywords[3].score | 0.5378889441490173 |
| keywords[3].display_name | Blood cell |
| keywords[4].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[4].score | 0.532296895980835 |
| keywords[4].display_name | Artificial intelligence |
| keywords[5].id | https://openalex.org/keywords/white-blood-cell |
| keywords[5].score | 0.514819324016571 |
| keywords[5].display_name | White blood cell |
| keywords[6].id | https://openalex.org/keywords/deep-learning |
| keywords[6].score | 0.5147202610969543 |
| keywords[6].display_name | Deep learning |
| keywords[7].id | https://openalex.org/keywords/human-blood |
| keywords[7].score | 0.47320979833602905 |
| keywords[7].display_name | Human blood |
| keywords[8].id | https://openalex.org/keywords/residual-neural-network |
| keywords[8].score | 0.4637741446495056 |
| keywords[8].display_name | Residual neural network |
| keywords[9].id | https://openalex.org/keywords/computer-science |
| keywords[9].score | 0.460194855928421 |
| keywords[9].display_name | Computer science |
| keywords[10].id | https://openalex.org/keywords/pattern-recognition |
| keywords[10].score | 0.4227150082588196 |
| keywords[10].display_name | Pattern recognition (psychology) |
| keywords[11].id | https://openalex.org/keywords/immunology |
| keywords[11].score | 0.3050536513328552 |
| keywords[11].display_name | Immunology |
| keywords[12].id | https://openalex.org/keywords/medicine |
| keywords[12].score | 0.278763085603714 |
| keywords[12].display_name | Medicine |
| language | en |
| locations[0].id | pmh:oai:arXiv.org:2308.06300 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400194 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | arXiv (Cornell University) |
| locations[0].source.host_organization | https://openalex.org/I205783295 |
| locations[0].source.host_organization_name | Cornell University |
| locations[0].source.host_organization_lineage | https://openalex.org/I205783295 |
| locations[0].license | |
| locations[0].pdf_url | https://arxiv.org/pdf/2308.06300 |
| locations[0].version | submittedVersion |
| locations[0].raw_type | text |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | http://arxiv.org/abs/2308.06300 |
| locations[1].id | doi:10.48550/arxiv.2308.06300 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400194 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | True |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | arXiv (Cornell University) |
| locations[1].source.host_organization | https://openalex.org/I205783295 |
| locations[1].source.host_organization_name | Cornell University |
| locations[1].source.host_organization_lineage | https://openalex.org/I205783295 |
| locations[1].license | cc-by |
| locations[1].pdf_url | |
| locations[1].version | |
| locations[1].raw_type | article-journal |
| locations[1].license_id | https://openalex.org/licenses/cc-by |
| locations[1].is_accepted | False |
| locations[1].is_published | |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://doi.org/10.48550/arxiv.2308.06300 |
| indexed_in | arxiv, datacite |
| authorships[0].author.id | https://openalex.org/A5092379594 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Rabia Asghar |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Asghar, Rabia |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5067847765 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-8951-5996 |
| authorships[1].author.display_name | Sanjay Kumar |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Kumar, Sanjay |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5092645122 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Abeera Mahfooz |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | Mahfooz, Abeera |
| authorships[2].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://arxiv.org/pdf/2308.06300 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2023-08-16T00:00:00 |
| display_name | Classification of All Blood Cell Images using ML and DL Models |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T12874 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1707 |
| primary_topic.subfield.display_name | Computer Vision and Pattern Recognition |
| primary_topic.display_name | Digital Imaging for Blood Diseases |
| related_works | https://openalex.org/W4205117943, https://openalex.org/W3179744494, https://openalex.org/W4362670567, https://openalex.org/W2882771868, https://openalex.org/W3202877052, https://openalex.org/W3162785402, https://openalex.org/W4224930320, https://openalex.org/W2371764668, https://openalex.org/W3032076087, https://openalex.org/W2518974813 |
| cited_by_count | 2 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 2 |
| locations_count | 2 |
| best_oa_location.id | pmh:oai:arXiv.org:2308.06300 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400194 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | arXiv (Cornell University) |
| best_oa_location.source.host_organization | https://openalex.org/I205783295 |
| best_oa_location.source.host_organization_name | Cornell University |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I205783295 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://arxiv.org/pdf/2308.06300 |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | text |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | http://arxiv.org/abs/2308.06300 |
| primary_location.id | pmh:oai:arXiv.org:2308.06300 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400194 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | arXiv (Cornell University) |
| primary_location.source.host_organization | https://openalex.org/I205783295 |
| primary_location.source.host_organization_name | Cornell University |
| primary_location.source.host_organization_lineage | https://openalex.org/I205783295 |
| primary_location.license | |
| primary_location.pdf_url | https://arxiv.org/pdf/2308.06300 |
| primary_location.version | submittedVersion |
| primary_location.raw_type | text |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | http://arxiv.org/abs/2308.06300 |
| publication_date | 2023-08-11 |
| publication_year | 2023 |
| referenced_works_count | 0 |
| abstract_inverted_index.A | 191 |
| abstract_inverted_index.a | 15, 106, 174 |
| abstract_inverted_index.In | 131 |
| abstract_inverted_index.It | 13 |
| abstract_inverted_index.an | 57, 230 |
| abstract_inverted_index.by | 170 |
| abstract_inverted_index.in | 18, 98, 252 |
| abstract_inverted_index.is | 115, 125 |
| abstract_inverted_index.it | 25, 124 |
| abstract_inverted_index.of | 88, 185, 216, 232 |
| abstract_inverted_index.on | 85, 208, 234 |
| abstract_inverted_index.or | 83 |
| abstract_inverted_index.to | 21, 38, 120, 127, 150, 179, 198, 248 |
| abstract_inverted_index.CNN | 137, 203 |
| abstract_inverted_index.Our | 238 |
| abstract_inverted_index.PBC | 152, 210, 236 |
| abstract_inverted_index.The | 112, 156, 201, 214 |
| abstract_inverted_index.and | 11, 48, 78, 81, 92, 96, 117, 147, 166 |
| abstract_inverted_index.are | 36 |
| abstract_inverted_index.can | 53 |
| abstract_inverted_index.for | 225 |
| abstract_inverted_index.has | 176, 195, 205 |
| abstract_inverted_index.our | 132, 221 |
| abstract_inverted_index.red | 5 |
| abstract_inverted_index.ten | 183 |
| abstract_inverted_index.the | 31, 40, 86, 151, 182, 209, 217, 235, 253 |
| abstract_inverted_index.DIB. | 155, 213 |
| abstract_inverted_index.been | 64, 177, 196, 206 |
| abstract_inverted_index.body | 41 |
| abstract_inverted_index.cell | 227 |
| abstract_inverted_index.data | 29 |
| abstract_inverted_index.have | 63, 100 |
| abstract_inverted_index.help | 54 |
| abstract_inverted_index.into | 66 |
| abstract_inverted_index.lies | 163 |
| abstract_inverted_index.more | 118 |
| abstract_inverted_index.role | 17 |
| abstract_inverted_index.that | 220 |
| abstract_inverted_index.this | 129 |
| abstract_inverted_index.when | 246 |
| abstract_inverted_index.with | 136, 160, 188 |
| abstract_inverted_index.Blood | 34, 61 |
| abstract_inverted_index.Human | 0 |
| abstract_inverted_index.about | 30 |
| abstract_inverted_index.basis | 87 |
| abstract_inverted_index.blood | 1, 6, 9, 51, 103, 186, 226 |
| abstract_inverted_index.body. | 33 |
| abstract_inverted_index.cells | 35, 62, 104, 187 |
| abstract_inverted_index.eight | 67 |
| abstract_inverted_index.human | 32, 121 |
| abstract_inverted_index.model | 175, 204, 243 |
| abstract_inverted_index.novel | 192 |
| abstract_inverted_index.plays | 14 |
| abstract_inverted_index.prone | 119 |
| abstract_inverted_index.their | 89 |
| abstract_inverted_index.them. | 111 |
| abstract_inverted_index.these | 102, 161, 171 |
| abstract_inverted_index.types | 184 |
| abstract_inverted_index.using | 105 |
| abstract_inverted_index.vital | 16 |
| abstract_inverted_index.where | 24 |
| abstract_inverted_index.white | 8 |
| abstract_inverted_index.91.375 | 165 |
| abstract_inverted_index.99.91% | 233 |
| abstract_inverted_index.Hence, | 50, 168 |
| abstract_inverted_index.VGG16, | 140 |
| abstract_inverted_index.VGG19, | 141 |
| abstract_inverted_index.assess | 56 |
| abstract_inverted_index.before | 108 |
| abstract_inverted_index.cells, | 7, 10 |
| abstract_inverted_index.defend | 39 |
| abstract_inverted_index.error. | 122 |
| abstract_inverted_index.fungi, | 46 |
| abstract_inverted_index.manual | 113 |
| abstract_inverted_index.models | 162 |
| abstract_inverted_index.neural | 241 |
| abstract_inverted_index.normal | 154, 212 |
| abstract_inverted_index.paper, | 133 |
| abstract_inverted_index.shape, | 91 |
| abstract_inverted_index.slower | 116 |
| abstract_inverted_index.stores | 26 |
| abstract_inverted_index.tested | 207 |
| abstract_inverted_index.94.72%. | 167 |
| abstract_inverted_index.against | 42 |
| abstract_inverted_index.applied | 149 |
| abstract_inverted_index.attains | 229 |
| abstract_inverted_index.between | 164 |
| abstract_inverted_index.dataset | 211 |
| abstract_inverted_index.diverse | 43 |
| abstract_inverted_index.earlier | 249 |
| abstract_inverted_index.groups: | 68 |
| abstract_inverted_index.improve | 199 |
| abstract_inverted_index.models. | 139 |
| abstract_inverted_index.network | 242 |
| abstract_inverted_index.organs, | 23 |
| abstract_inverted_index.overall | 157 |
| abstract_inverted_index.plasma, | 4 |
| abstract_inverted_index.results | 250 |
| abstract_inverted_index.accuracy | 158, 231 |
| abstract_inverted_index.achieved | 159 |
| abstract_inverted_index.analysis | 52 |
| abstract_inverted_index.approach | 114 |
| abstract_inverted_index.automate | 128 |
| abstract_inverted_index.classify | 181 |
| abstract_inverted_index.compared | 247 |
| abstract_inverted_index.dataset. | 237 |
| abstract_inverted_index.designed | 224 |
| abstract_inverted_index.examined | 101 |
| abstract_inverted_index.immature | 74 |
| abstract_inverted_index.inspired | 169 |
| abstract_inverted_index.learning | 135 |
| abstract_inverted_index.manually | 109 |
| abstract_inverted_index.nucleus, | 90 |
| abstract_inverted_index.outcomes | 215 |
| abstract_inverted_index.performs | 244 |
| abstract_inverted_index.process. | 130 |
| abstract_inverted_index.proposed | 178, 202, 239 |
| abstract_inverted_index.reported | 251 |
| abstract_inverted_index.transfer | 134 |
| abstract_inverted_index.utilized | 37 |
| abstract_inverted_index.viruses, | 47 |
| abstract_inverted_index.CNN-based | 193, 222 |
| abstract_inverted_index.accuracy. | 190, 200 |
| abstract_inverted_index.bacteria. | 49 |
| abstract_inverted_index.comprises | 3 |
| abstract_inverted_index.dataset's | 153 |
| abstract_inverted_index.different | 22 |
| abstract_inverted_index.essential | 27, 126 |
| abstract_inverted_index.framework | 194, 223 |
| abstract_inverted_index.including | 45 |
| abstract_inverted_index.increased | 189 |
| abstract_inverted_index.nutrients | 20 |
| abstract_inverted_index.platelets | 82 |
| abstract_inverted_index.presented | 197 |
| abstract_inverted_index.primarily | 2 |
| abstract_inverted_index.ResNet-50, | 142 |
| abstract_inverted_index.Therefore, | 123 |
| abstract_inverted_index.basophils, | 71 |
| abstract_inverted_index.condition. | 60 |
| abstract_inverted_index.cytoplasm. | 93 |
| abstract_inverted_index.microscope | 107 |
| abstract_inverted_index.monocytes, | 73 |
| abstract_inverted_index.physicians | 55 |
| abstract_inverted_index.platelets. | 12 |
| abstract_inverted_index.DenseNet-20 | 148 |
| abstract_inverted_index.ResNet-101, | 143 |
| abstract_inverted_index.ResNet-152, | 144 |
| abstract_inverted_index.classifying | 110 |
| abstract_inverted_index.demonstrate | 219 |
| abstract_inverted_index.experiments | 218 |
| abstract_inverted_index.infections, | 44 |
| abstract_inverted_index.literature. | 254 |
| abstract_inverted_index.myelocytes, | 77 |
| abstract_inverted_index.pre-trained | 138, 172 |
| abstract_inverted_index.InceptionV3, | 145 |
| abstract_inverted_index.MobileNetV2, | 146 |
| abstract_inverted_index.Neutrophils, | 69 |
| abstract_inverted_index.eosinophils, | 70 |
| abstract_inverted_index.granulocytes | 75 |
| abstract_inverted_index.individual's | 58 |
| abstract_inverted_index.laboratories | 99 |
| abstract_inverted_index.lymphocytes, | 72 |
| abstract_inverted_index.pathologists | 95 |
| abstract_inverted_index.thrombocytes | 84 |
| abstract_inverted_index.transporting | 19 |
| abstract_inverted_index.automatically | 180 |
| abstract_inverted_index.competitively | 245 |
| abstract_inverted_index.convolutional | 240 |
| abstract_inverted_index.hematologists | 97 |
| abstract_inverted_index.physiological | 59 |
| abstract_inverted_index.Traditionally, | 94 |
| abstract_inverted_index.architectures, | 173 |
| abstract_inverted_index.classification | 228 |
| abstract_inverted_index.erythroblasts, | 80 |
| abstract_inverted_index.health-related | 28 |
| abstract_inverted_index.sub-classified | 65 |
| abstract_inverted_index.(promyelocytes, | 76 |
| abstract_inverted_index.metamyelocytes), | 79 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 95 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 3 |
| citation_normalized_percentile.value | 0.55839174 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |