Classification of RNA Pseudoknots and Comparison of Structure Prediction Methods Article Swipe
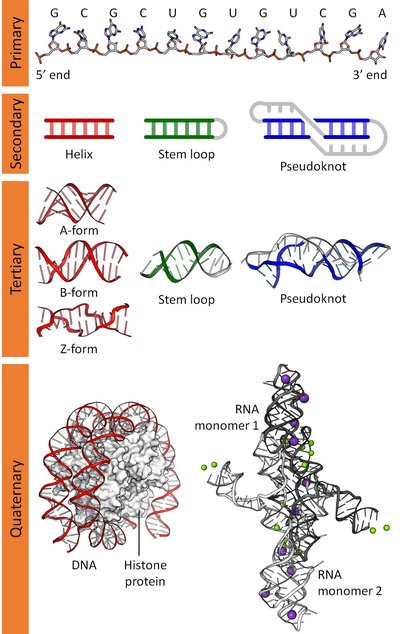
Lots of researches convey the importance of the RNA molecules, as they play vital roles in many molecular procedures. And it is commonly believed that the structures of the RNA molecules hold the key to the discovery of their functions.During the investigation of RNA structures, the researchers are dependent on the bioinformatical methods increasingly. Many in silico methods of predicting RNA secondary structures have emerged in this big wave, including some ones which are capable of predicting pseudoknots, a particular type of RNA secondary structures.The purpose of this dissertation is to try to compare the state-of-the-art methods predicting pseudoknots, and offer the colleagues some insights into how to choose a practical method for the given single sequence. In fact, lots of efforts have been done into the prediction of RNA secondary structures including pseudoknots during the last decades, contributing to many programs in this field. Some challenging questions are raised consequently. How about the performance of each method, especially on a particular class of RNA sequences? What are their advantages and disadvantages? What can we benefit from the contemporary methods if we want to develop new ones? This dissertation holds the confidence in the investigation of the answers.This dissertation carries out quite many comparisons of the performance of predicting RNA pseudoknots by the available methods. One main part focuses on the prediction of frameshifting signals by two methods principally. The second main part focuses on the prediction of pseudoknots which participate in much more general molecular activities.In detail, the second part of work includes 414 pseudoknots, from both the Pseudobase and the Protein Data Bank, and 15 methods including 3 exact methods and 12 heuristic ones. Specifically, three main categories of complexity measurements are introduced, which further divide the 414 pseudoknots into a series of subclasses respectively. The comparisons are carried out by comparing the predictions of each method based on the entire 414 pseudoknots, and the subsets which are classified by both the complexity measurements and the length, RNA type and organism of the pseudoknots.The result shows that the pseudoknots in nature hold a relatively low complexity in all measurements. And the performance of contemporary methods varies from subclass to subclass, but decreases consistently as the complexity of pseudoknots increases. More generally, the heuristic methods globally outperform the exact ones. And the susceptible assessment results are dependent strongly on the quality of the reference structures and the evaluation system. Last but not least, this part of work is provided as an on-line benchmark for the bioinformatics community.
Related Topics
- Type
- preprint
- Language
- fr
- Landing Page
- https://tel.archives-ouvertes.fr/tel-01297053
- OA Status
- green
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2247317260
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2247317260Canonical identifier for this work in OpenAlex
- Title
-
Classification of RNA Pseudoknots and Comparison of Structure Prediction MethodsWork title
- Type
-
preprintOpenAlex work type
- Language
-
frPrimary language
- Publication year
-
2015Year of publication
- Publication date
-
2015-07-03Full publication date if available
- Authors
-
Cong ZengList of authors in order
- Landing page
-
https://tel.archives-ouvertes.fr/tel-01297053Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://tel.archives-ouvertes.fr/tel-01297053Direct OA link when available
- Concepts
-
Nucleic acid secondary structure, RNA, Computer science, Computational biology, Pseudoknot, Nucleic acid structure, In silico, Data science, Biology, Genetics, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2247317260 |
|---|---|
| doi | |
| ids.mag | 2247317260 |
| ids.openalex | https://openalex.org/W2247317260 |
| fwci | |
| type | preprint |
| title | Classification of RNA Pseudoknots and Comparison of Structure Prediction Methods |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10521 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9986000061035156 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | RNA and protein synthesis mechanisms |
| topics[1].id | https://openalex.org/T12254 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9898999929428101 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Machine Learning in Bioinformatics |
| topics[2].id | https://openalex.org/T10044 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9513000249862671 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Protein Structure and Dynamics |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C85468374 |
| concepts[0].level | 4 |
| concepts[0].score | 0.5739542841911316 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q6589957 |
| concepts[0].display_name | Nucleic acid secondary structure |
| concepts[1].id | https://openalex.org/C67705224 |
| concepts[1].level | 3 |
| concepts[1].score | 0.5723136067390442 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q11053 |
| concepts[1].display_name | RNA |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.5581456422805786 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C70721500 |
| concepts[3].level | 1 |
| concepts[3].score | 0.5196424722671509 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[3].display_name | Computational biology |
| concepts[4].id | https://openalex.org/C105130999 |
| concepts[4].level | 4 |
| concepts[4].score | 0.4816421568393707 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q414976 |
| concepts[4].display_name | Pseudoknot |
| concepts[5].id | https://openalex.org/C126142528 |
| concepts[5].level | 4 |
| concepts[5].score | 0.4670174717903137 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q3771876 |
| concepts[5].display_name | Nucleic acid structure |
| concepts[6].id | https://openalex.org/C2775905019 |
| concepts[6].level | 3 |
| concepts[6].score | 0.42691484093666077 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q192572 |
| concepts[6].display_name | In silico |
| concepts[7].id | https://openalex.org/C2522767166 |
| concepts[7].level | 1 |
| concepts[7].score | 0.368264377117157 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q2374463 |
| concepts[7].display_name | Data science |
| concepts[8].id | https://openalex.org/C86803240 |
| concepts[8].level | 0 |
| concepts[8].score | 0.24801838397979736 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[8].display_name | Biology |
| concepts[9].id | https://openalex.org/C54355233 |
| concepts[9].level | 1 |
| concepts[9].score | 0.16333931684494019 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[9].display_name | Genetics |
| concepts[10].id | https://openalex.org/C104317684 |
| concepts[10].level | 2 |
| concepts[10].score | 0.09044808149337769 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[10].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/nucleic-acid-secondary-structure |
| keywords[0].score | 0.5739542841911316 |
| keywords[0].display_name | Nucleic acid secondary structure |
| keywords[1].id | https://openalex.org/keywords/rna |
| keywords[1].score | 0.5723136067390442 |
| keywords[1].display_name | RNA |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.5581456422805786 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/computational-biology |
| keywords[3].score | 0.5196424722671509 |
| keywords[3].display_name | Computational biology |
| keywords[4].id | https://openalex.org/keywords/pseudoknot |
| keywords[4].score | 0.4816421568393707 |
| keywords[4].display_name | Pseudoknot |
| keywords[5].id | https://openalex.org/keywords/nucleic-acid-structure |
| keywords[5].score | 0.4670174717903137 |
| keywords[5].display_name | Nucleic acid structure |
| keywords[6].id | https://openalex.org/keywords/in-silico |
| keywords[6].score | 0.42691484093666077 |
| keywords[6].display_name | In silico |
| keywords[7].id | https://openalex.org/keywords/data-science |
| keywords[7].score | 0.368264377117157 |
| keywords[7].display_name | Data science |
| keywords[8].id | https://openalex.org/keywords/biology |
| keywords[8].score | 0.24801838397979736 |
| keywords[8].display_name | Biology |
| keywords[9].id | https://openalex.org/keywords/genetics |
| keywords[9].score | 0.16333931684494019 |
| keywords[9].display_name | Genetics |
| keywords[10].id | https://openalex.org/keywords/gene |
| keywords[10].score | 0.09044808149337769 |
| keywords[10].display_name | Gene |
| language | fr |
| locations[0].id | mag:2247317260 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402512 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| locations[0].source.host_organization | https://openalex.org/I1294671590 |
| locations[0].source.host_organization_name | Centre National de la Recherche Scientifique |
| locations[0].source.host_organization_lineage | https://openalex.org/I1294671590 |
| locations[0].license | other-oa |
| locations[0].pdf_url | |
| locations[0].version | submittedVersion |
| locations[0].raw_type | |
| locations[0].license_id | https://openalex.org/licenses/other-oa |
| locations[0].is_accepted | False |
| locations[0].is_published | False |
| locations[0].raw_source_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| locations[0].landing_page_url | https://tel.archives-ouvertes.fr/tel-01297053 |
| locations[1].id | pmh:2015PA112127 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400553 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | Munich Personal RePEc Archive (Ludwig Maximilian University of Munich) |
| locations[1].source.host_organization | https://openalex.org/I8204097 |
| locations[1].source.host_organization_name | Ludwig-Maximilians-Universität München |
| locations[1].source.host_organization_lineage | https://openalex.org/I8204097 |
| locations[1].license | other-oa |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Electronic Thesis or Dissertation |
| locations[1].license_id | https://openalex.org/licenses/other-oa |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | http://www.theses.fr/2015PA112127/document |
| locations[2].id | pmh:oai:HAL:tel-01297053v1 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306402512 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| locations[2].source.host_organization | https://openalex.org/I1294671590 |
| locations[2].source.host_organization_name | Centre National de la Recherche Scientifique |
| locations[2].source.host_organization_lineage | https://openalex.org/I1294671590 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Theses |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Bioinformatics [q-bio.QM]. Université Paris Sud - Paris XI, 2015. English. ⟨NNT : 2015PA112127⟩ |
| locations[2].landing_page_url | https://theses.hal.science/tel-01297053 |
| authorships[0].author.id | https://openalex.org/A5101701544 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-9282-5549 |
| authorships[0].author.display_name | Cong Zeng |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Zeng, Cong |
| authorships[0].is_corresponding | True |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://tel.archives-ouvertes.fr/tel-01297053 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Classification of RNA Pseudoknots and Comparison of Structure Prediction Methods |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-10-10T17:16:08.811792 |
| primary_topic.id | https://openalex.org/T10521 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9986000061035156 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | RNA and protein synthesis mechanisms |
| related_works | https://openalex.org/W2159621058, https://openalex.org/W2954092143, https://openalex.org/W2148725278, https://openalex.org/W1583489351, https://openalex.org/W2135373808, https://openalex.org/W2105081912, https://openalex.org/W2796011749, https://openalex.org/W1997175692, https://openalex.org/W2018280025, https://openalex.org/W2740179092 |
| cited_by_count | 0 |
| locations_count | 3 |
| best_oa_location.id | mag:2247317260 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402512 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| best_oa_location.source.host_organization | https://openalex.org/I1294671590 |
| best_oa_location.source.host_organization_name | Centre National de la Recherche Scientifique |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I1294671590 |
| best_oa_location.license | other-oa |
| best_oa_location.pdf_url | |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | |
| best_oa_location.license_id | https://openalex.org/licenses/other-oa |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| best_oa_location.landing_page_url | https://tel.archives-ouvertes.fr/tel-01297053 |
| primary_location.id | mag:2247317260 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402512 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| primary_location.source.host_organization | https://openalex.org/I1294671590 |
| primary_location.source.host_organization_name | Centre National de la Recherche Scientifique |
| primary_location.source.host_organization_lineage | https://openalex.org/I1294671590 |
| primary_location.license | other-oa |
| primary_location.pdf_url | |
| primary_location.version | submittedVersion |
| primary_location.raw_type | |
| primary_location.license_id | https://openalex.org/licenses/other-oa |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| primary_location.landing_page_url | https://tel.archives-ouvertes.fr/tel-01297053 |
| publication_date | 2015-07-03 |
| publication_year | 2015 |
| referenced_works_count | 0 |
| abstract_inverted_index.3 | 269 |
| abstract_inverted_index.a | 78, 109, 160, 292, 344 |
| abstract_inverted_index.12 | 273 |
| abstract_inverted_index.15 | 266 |
| abstract_inverted_index.In | 117 |
| abstract_inverted_index.an | 411 |
| abstract_inverted_index.as | 10, 365, 410 |
| abstract_inverted_index.by | 211, 225, 302, 321 |
| abstract_inverted_index.if | 180 |
| abstract_inverted_index.in | 15, 55, 65, 142, 192, 241, 341, 348 |
| abstract_inverted_index.is | 21, 89, 408 |
| abstract_inverted_index.it | 20 |
| abstract_inverted_index.of | 1, 6, 27, 37, 42, 58, 75, 81, 86, 120, 128, 155, 163, 195, 204, 207, 222, 237, 251, 280, 294, 306, 333, 354, 368, 392, 406 |
| abstract_inverted_index.on | 49, 159, 219, 234, 310, 389 |
| abstract_inverted_index.to | 34, 90, 92, 107, 139, 183, 360 |
| abstract_inverted_index.we | 174, 181 |
| abstract_inverted_index.414 | 254, 289, 313 |
| abstract_inverted_index.And | 19, 351, 381 |
| abstract_inverted_index.How | 151 |
| abstract_inverted_index.One | 215 |
| abstract_inverted_index.RNA | 8, 29, 43, 60, 82, 129, 164, 209, 329 |
| abstract_inverted_index.The | 229, 297 |
| abstract_inverted_index.all | 349 |
| abstract_inverted_index.and | 99, 170, 260, 265, 272, 315, 326, 331, 396 |
| abstract_inverted_index.are | 47, 73, 148, 167, 283, 299, 319, 386 |
| abstract_inverted_index.big | 67 |
| abstract_inverted_index.but | 362, 401 |
| abstract_inverted_index.can | 173 |
| abstract_inverted_index.for | 112, 414 |
| abstract_inverted_index.how | 106 |
| abstract_inverted_index.key | 33 |
| abstract_inverted_index.low | 346 |
| abstract_inverted_index.new | 185 |
| abstract_inverted_index.not | 402 |
| abstract_inverted_index.out | 200, 301 |
| abstract_inverted_index.the | 4, 7, 25, 28, 32, 35, 40, 45, 50, 94, 101, 113, 126, 135, 153, 177, 190, 193, 196, 205, 212, 220, 235, 248, 258, 261, 288, 304, 311, 316, 323, 327, 334, 339, 352, 366, 373, 378, 382, 390, 393, 397, 415 |
| abstract_inverted_index.try | 91 |
| abstract_inverted_index.two | 226 |
| abstract_inverted_index.Data | 263 |
| abstract_inverted_index.Last | 400 |
| abstract_inverted_index.Lots | 0 |
| abstract_inverted_index.Many | 54 |
| abstract_inverted_index.More | 371 |
| abstract_inverted_index.Some | 145 |
| abstract_inverted_index.This | 187 |
| abstract_inverted_index.What | 166, 172 |
| abstract_inverted_index.been | 123 |
| abstract_inverted_index.both | 257, 322 |
| abstract_inverted_index.done | 124 |
| abstract_inverted_index.each | 156, 307 |
| abstract_inverted_index.from | 176, 256, 358 |
| abstract_inverted_index.have | 63, 122 |
| abstract_inverted_index.hold | 31, 343 |
| abstract_inverted_index.into | 105, 125, 291 |
| abstract_inverted_index.last | 136 |
| abstract_inverted_index.lots | 119 |
| abstract_inverted_index.main | 216, 231, 278 |
| abstract_inverted_index.many | 16, 140, 202 |
| abstract_inverted_index.more | 243 |
| abstract_inverted_index.much | 242 |
| abstract_inverted_index.ones | 71 |
| abstract_inverted_index.part | 217, 232, 250, 405 |
| abstract_inverted_index.play | 12 |
| abstract_inverted_index.some | 70, 103 |
| abstract_inverted_index.that | 24, 338 |
| abstract_inverted_index.they | 11 |
| abstract_inverted_index.this | 66, 87, 143, 404 |
| abstract_inverted_index.type | 80, 330 |
| abstract_inverted_index.want | 182 |
| abstract_inverted_index.work | 252, 407 |
| abstract_inverted_index.Bank, | 264 |
| abstract_inverted_index.about | 152 |
| abstract_inverted_index.based | 309 |
| abstract_inverted_index.class | 162 |
| abstract_inverted_index.exact | 270, 379 |
| abstract_inverted_index.fact, | 118 |
| abstract_inverted_index.given | 114 |
| abstract_inverted_index.holds | 189 |
| abstract_inverted_index.offer | 100 |
| abstract_inverted_index.ones. | 275, 380 |
| abstract_inverted_index.ones? | 186 |
| abstract_inverted_index.quite | 201 |
| abstract_inverted_index.roles | 14 |
| abstract_inverted_index.shows | 337 |
| abstract_inverted_index.their | 38, 168 |
| abstract_inverted_index.three | 277 |
| abstract_inverted_index.vital | 13 |
| abstract_inverted_index.wave, | 68 |
| abstract_inverted_index.which | 72, 239, 285, 318 |
| abstract_inverted_index.choose | 108 |
| abstract_inverted_index.convey | 3 |
| abstract_inverted_index.divide | 287 |
| abstract_inverted_index.during | 134 |
| abstract_inverted_index.entire | 312 |
| abstract_inverted_index.field. | 144 |
| abstract_inverted_index.least, | 403 |
| abstract_inverted_index.method | 111, 308 |
| abstract_inverted_index.nature | 342 |
| abstract_inverted_index.raised | 149 |
| abstract_inverted_index.result | 336 |
| abstract_inverted_index.second | 230, 249 |
| abstract_inverted_index.series | 293 |
| abstract_inverted_index.silico | 56 |
| abstract_inverted_index.single | 115 |
| abstract_inverted_index.varies | 357 |
| abstract_inverted_index.Protein | 262 |
| abstract_inverted_index.benefit | 175 |
| abstract_inverted_index.capable | 74 |
| abstract_inverted_index.carried | 300 |
| abstract_inverted_index.carries | 199 |
| abstract_inverted_index.compare | 93 |
| abstract_inverted_index.detail, | 247 |
| abstract_inverted_index.develop | 184 |
| abstract_inverted_index.efforts | 121 |
| abstract_inverted_index.emerged | 64 |
| abstract_inverted_index.focuses | 218, 233 |
| abstract_inverted_index.further | 286 |
| abstract_inverted_index.general | 244 |
| abstract_inverted_index.length, | 328 |
| abstract_inverted_index.method, | 157 |
| abstract_inverted_index.methods | 52, 57, 96, 179, 227, 267, 271, 356, 375 |
| abstract_inverted_index.on-line | 412 |
| abstract_inverted_index.purpose | 85 |
| abstract_inverted_index.quality | 391 |
| abstract_inverted_index.results | 385 |
| abstract_inverted_index.signals | 224 |
| abstract_inverted_index.subsets | 317 |
| abstract_inverted_index.system. | 399 |
| abstract_inverted_index.believed | 23 |
| abstract_inverted_index.commonly | 22 |
| abstract_inverted_index.decades, | 137 |
| abstract_inverted_index.globally | 376 |
| abstract_inverted_index.includes | 253 |
| abstract_inverted_index.insights | 104 |
| abstract_inverted_index.methods. | 214 |
| abstract_inverted_index.organism | 332 |
| abstract_inverted_index.programs | 141 |
| abstract_inverted_index.provided | 409 |
| abstract_inverted_index.strongly | 388 |
| abstract_inverted_index.subclass | 359 |
| abstract_inverted_index.available | 213 |
| abstract_inverted_index.benchmark | 413 |
| abstract_inverted_index.comparing | 303 |
| abstract_inverted_index.decreases | 363 |
| abstract_inverted_index.dependent | 48, 387 |
| abstract_inverted_index.discovery | 36 |
| abstract_inverted_index.heuristic | 274, 374 |
| abstract_inverted_index.including | 69, 132, 268 |
| abstract_inverted_index.molecular | 17, 245 |
| abstract_inverted_index.molecules | 30 |
| abstract_inverted_index.practical | 110 |
| abstract_inverted_index.questions | 147 |
| abstract_inverted_index.reference | 394 |
| abstract_inverted_index.secondary | 61, 83, 130 |
| abstract_inverted_index.sequence. | 116 |
| abstract_inverted_index.subclass, | 361 |
| abstract_inverted_index.Pseudobase | 259 |
| abstract_inverted_index.advantages | 169 |
| abstract_inverted_index.assessment | 384 |
| abstract_inverted_index.categories | 279 |
| abstract_inverted_index.classified | 320 |
| abstract_inverted_index.colleagues | 102 |
| abstract_inverted_index.community. | 417 |
| abstract_inverted_index.complexity | 281, 324, 347, 367 |
| abstract_inverted_index.confidence | 191 |
| abstract_inverted_index.especially | 158 |
| abstract_inverted_index.evaluation | 398 |
| abstract_inverted_index.generally, | 372 |
| abstract_inverted_index.importance | 5 |
| abstract_inverted_index.increases. | 370 |
| abstract_inverted_index.molecules, | 9 |
| abstract_inverted_index.outperform | 377 |
| abstract_inverted_index.particular | 79, 161 |
| abstract_inverted_index.predicting | 59, 76, 97, 208 |
| abstract_inverted_index.prediction | 127, 221, 236 |
| abstract_inverted_index.relatively | 345 |
| abstract_inverted_index.researches | 2 |
| abstract_inverted_index.sequences? | 165 |
| abstract_inverted_index.structures | 26, 62, 131, 395 |
| abstract_inverted_index.subclasses | 295 |
| abstract_inverted_index.challenging | 146 |
| abstract_inverted_index.comparisons | 203, 298 |
| abstract_inverted_index.introduced, | 284 |
| abstract_inverted_index.participate | 240 |
| abstract_inverted_index.performance | 154, 206, 353 |
| abstract_inverted_index.predictions | 305 |
| abstract_inverted_index.procedures. | 18 |
| abstract_inverted_index.pseudoknots | 133, 210, 238, 290, 340, 369 |
| abstract_inverted_index.researchers | 46 |
| abstract_inverted_index.structures, | 44 |
| abstract_inverted_index.susceptible | 383 |
| abstract_inverted_index.answers.This | 197 |
| abstract_inverted_index.consistently | 364 |
| abstract_inverted_index.contemporary | 178, 355 |
| abstract_inverted_index.contributing | 138 |
| abstract_inverted_index.dissertation | 88, 188, 198 |
| abstract_inverted_index.measurements | 282, 325 |
| abstract_inverted_index.principally. | 228 |
| abstract_inverted_index.pseudoknots, | 77, 98, 255, 314 |
| abstract_inverted_index.Specifically, | 276 |
| abstract_inverted_index.activities.In | 246 |
| abstract_inverted_index.consequently. | 150 |
| abstract_inverted_index.frameshifting | 223 |
| abstract_inverted_index.increasingly. | 53 |
| abstract_inverted_index.investigation | 41, 194 |
| abstract_inverted_index.measurements. | 350 |
| abstract_inverted_index.respectively. | 296 |
| abstract_inverted_index.bioinformatics | 416 |
| abstract_inverted_index.disadvantages? | 171 |
| abstract_inverted_index.structures.The | 84 |
| abstract_inverted_index.bioinformatical | 51 |
| abstract_inverted_index.pseudoknots.The | 335 |
| abstract_inverted_index.functions.During | 39 |
| abstract_inverted_index.state-of-the-art | 95 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5101701544 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 1 |
| citation_normalized_percentile |