Comprehensive Guide of Epigenetics and Transcriptomics Data Quality Control Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.08.02.606411
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1101/2024.08.02.606411
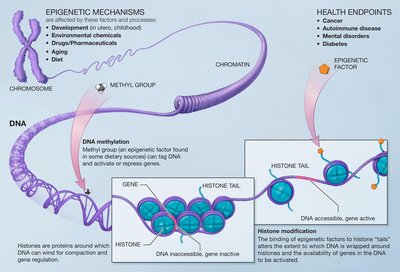
Host response to environmental exposures such as pathogens and chemicals can cause modifications to the epigenome and transcriptome. Analysis of these modifications can reveal signatures with regards to the agent and timing of exposure. Exhaustive interrogation of the cascade of the epigenome and transcriptome requires analysis of disparate datasets from multiple assay types, often at single cell resolution, from the same biospecimen. Improved signature discovery has been enabled by advancements in assaying techniques to detect RNA expression, DNA base modifications, histone modifications, and chromatin accessibility. However, there remains a paucity of rigorous quality control standards of those datasets that reflect quality assurance of the underlying assay. This guide outlines a comprehensive suite of metrics that can be used to ensure quality from 11 different epigenetics and transcriptomics assays. Recommendations on mitigation approaches to address failed metrics and poor quality data are provided. The workflow consists of assessing dataset quality and reiterating benchwork protocols for improved results to generate accurate exposure signatures.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2024.08.02.606411
- https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdf
- OA Status
- green
- References
- 71
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4401406573
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4401406573Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2024.08.02.606411Digital Object Identifier
- Title
-
Comprehensive Guide of Epigenetics and Transcriptomics Data Quality ControlWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-08-06Full publication date if available
- Authors
-
Arianna Comendul, Frederique Ruf-Zamojski, Colby T. Ford, Pankaj Agarwal, Elena Zaslavsky, German Nudelman, Manoj Hariharan, Aliza B. Rubenstein, Hanna Pinças, Venugopalan D. Nair, Adam Michaleas, Stuart C. Sealfon, Christopher W. Woods, Kajal Claypool, Rafael JaimesList of authors in order
- Landing page
-
https://doi.org/10.1101/2024.08.02.606411Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdfDirect OA link when available
- Concepts
-
Epigenome, Workflow, Computational biology, Computer science, Quality (philosophy), Epigenetics, Epigenomics, Transcriptome, Histone, Chromatin, Data mining, Data science, Biology, DNA methylation, Genetics, Gene expression, DNA, Gene, Database, Epistemology, PhilosophyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
71Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4401406573 |
|---|---|
| doi | https://doi.org/10.1101/2024.08.02.606411 |
| ids.doi | https://doi.org/10.1101/2024.08.02.606411 |
| ids.openalex | https://openalex.org/W4401406573 |
| fwci | 0.0 |
| type | preprint |
| title | Comprehensive Guide of Epigenetics and Transcriptomics Data Quality Control |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10269 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9991999864578247 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Epigenetics and DNA Methylation |
| topics[1].id | https://openalex.org/T11482 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9990000128746033 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | RNA modifications and cancer |
| topics[2].id | https://openalex.org/T11289 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9983999729156494 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Single-cell and spatial transcriptomics |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C117717151 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7731798887252808 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q3589155 |
| concepts[0].display_name | Epigenome |
| concepts[1].id | https://openalex.org/C177212765 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6329329013824463 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q627335 |
| concepts[1].display_name | Workflow |
| concepts[2].id | https://openalex.org/C70721500 |
| concepts[2].level | 1 |
| concepts[2].score | 0.6162639856338501 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[2].display_name | Computational biology |
| concepts[3].id | https://openalex.org/C41008148 |
| concepts[3].level | 0 |
| concepts[3].score | 0.5916886925697327 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[3].display_name | Computer science |
| concepts[4].id | https://openalex.org/C2779530757 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5163958668708801 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1207505 |
| concepts[4].display_name | Quality (philosophy) |
| concepts[5].id | https://openalex.org/C41091548 |
| concepts[5].level | 3 |
| concepts[5].score | 0.49767735600471497 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q26939 |
| concepts[5].display_name | Epigenetics |
| concepts[6].id | https://openalex.org/C121912465 |
| concepts[6].level | 5 |
| concepts[6].score | 0.45139122009277344 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q3589153 |
| concepts[6].display_name | Epigenomics |
| concepts[7].id | https://openalex.org/C162317418 |
| concepts[7].level | 4 |
| concepts[7].score | 0.4382012188434601 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q252857 |
| concepts[7].display_name | Transcriptome |
| concepts[8].id | https://openalex.org/C64927066 |
| concepts[8].level | 3 |
| concepts[8].score | 0.43731045722961426 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q36293 |
| concepts[8].display_name | Histone |
| concepts[9].id | https://openalex.org/C83640560 |
| concepts[9].level | 3 |
| concepts[9].score | 0.41790956258773804 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q180951 |
| concepts[9].display_name | Chromatin |
| concepts[10].id | https://openalex.org/C124101348 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3815075755119324 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q172491 |
| concepts[10].display_name | Data mining |
| concepts[11].id | https://openalex.org/C2522767166 |
| concepts[11].level | 1 |
| concepts[11].score | 0.36869046092033386 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q2374463 |
| concepts[11].display_name | Data science |
| concepts[12].id | https://openalex.org/C86803240 |
| concepts[12].level | 0 |
| concepts[12].score | 0.36836618185043335 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[12].display_name | Biology |
| concepts[13].id | https://openalex.org/C190727270 |
| concepts[13].level | 4 |
| concepts[13].score | 0.26058363914489746 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q874745 |
| concepts[13].display_name | DNA methylation |
| concepts[14].id | https://openalex.org/C54355233 |
| concepts[14].level | 1 |
| concepts[14].score | 0.14413002133369446 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[14].display_name | Genetics |
| concepts[15].id | https://openalex.org/C150194340 |
| concepts[15].level | 3 |
| concepts[15].score | 0.11950159072875977 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[15].display_name | Gene expression |
| concepts[16].id | https://openalex.org/C552990157 |
| concepts[16].level | 2 |
| concepts[16].score | 0.1118655800819397 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[16].display_name | DNA |
| concepts[17].id | https://openalex.org/C104317684 |
| concepts[17].level | 2 |
| concepts[17].score | 0.09267181158065796 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[17].display_name | Gene |
| concepts[18].id | https://openalex.org/C77088390 |
| concepts[18].level | 1 |
| concepts[18].score | 0.0901915431022644 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q8513 |
| concepts[18].display_name | Database |
| concepts[19].id | https://openalex.org/C111472728 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q9471 |
| concepts[19].display_name | Epistemology |
| concepts[20].id | https://openalex.org/C138885662 |
| concepts[20].level | 0 |
| concepts[20].score | 0.0 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q5891 |
| concepts[20].display_name | Philosophy |
| keywords[0].id | https://openalex.org/keywords/epigenome |
| keywords[0].score | 0.7731798887252808 |
| keywords[0].display_name | Epigenome |
| keywords[1].id | https://openalex.org/keywords/workflow |
| keywords[1].score | 0.6329329013824463 |
| keywords[1].display_name | Workflow |
| keywords[2].id | https://openalex.org/keywords/computational-biology |
| keywords[2].score | 0.6162639856338501 |
| keywords[2].display_name | Computational biology |
| keywords[3].id | https://openalex.org/keywords/computer-science |
| keywords[3].score | 0.5916886925697327 |
| keywords[3].display_name | Computer science |
| keywords[4].id | https://openalex.org/keywords/quality |
| keywords[4].score | 0.5163958668708801 |
| keywords[4].display_name | Quality (philosophy) |
| keywords[5].id | https://openalex.org/keywords/epigenetics |
| keywords[5].score | 0.49767735600471497 |
| keywords[5].display_name | Epigenetics |
| keywords[6].id | https://openalex.org/keywords/epigenomics |
| keywords[6].score | 0.45139122009277344 |
| keywords[6].display_name | Epigenomics |
| keywords[7].id | https://openalex.org/keywords/transcriptome |
| keywords[7].score | 0.4382012188434601 |
| keywords[7].display_name | Transcriptome |
| keywords[8].id | https://openalex.org/keywords/histone |
| keywords[8].score | 0.43731045722961426 |
| keywords[8].display_name | Histone |
| keywords[9].id | https://openalex.org/keywords/chromatin |
| keywords[9].score | 0.41790956258773804 |
| keywords[9].display_name | Chromatin |
| keywords[10].id | https://openalex.org/keywords/data-mining |
| keywords[10].score | 0.3815075755119324 |
| keywords[10].display_name | Data mining |
| keywords[11].id | https://openalex.org/keywords/data-science |
| keywords[11].score | 0.36869046092033386 |
| keywords[11].display_name | Data science |
| keywords[12].id | https://openalex.org/keywords/biology |
| keywords[12].score | 0.36836618185043335 |
| keywords[12].display_name | Biology |
| keywords[13].id | https://openalex.org/keywords/dna-methylation |
| keywords[13].score | 0.26058363914489746 |
| keywords[13].display_name | DNA methylation |
| keywords[14].id | https://openalex.org/keywords/genetics |
| keywords[14].score | 0.14413002133369446 |
| keywords[14].display_name | Genetics |
| keywords[15].id | https://openalex.org/keywords/gene-expression |
| keywords[15].score | 0.11950159072875977 |
| keywords[15].display_name | Gene expression |
| keywords[16].id | https://openalex.org/keywords/dna |
| keywords[16].score | 0.1118655800819397 |
| keywords[16].display_name | DNA |
| keywords[17].id | https://openalex.org/keywords/gene |
| keywords[17].score | 0.09267181158065796 |
| keywords[17].display_name | Gene |
| keywords[18].id | https://openalex.org/keywords/database |
| keywords[18].score | 0.0901915431022644 |
| keywords[18].display_name | Database |
| language | en |
| locations[0].id | doi:10.1101/2024.08.02.606411 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2024.08.02.606411 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5106354102 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Arianna Comendul |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[0].affiliations[0].raw_affiliation_string | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[0].institutions[0].id | https://openalex.org/I4210122954 |
| authorships[0].institutions[0].ror | https://ror.org/022z6jk58 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | MIT Lincoln Laboratory |
| authorships[0].institutions[1].id | https://openalex.org/I63966007 |
| authorships[0].institutions[1].ror | https://ror.org/042nb2s44 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I63966007 |
| authorships[0].institutions[1].country_code | US |
| authorships[0].institutions[1].display_name | Massachusetts Institute of Technology |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Arianna Comendul |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[1].author.id | https://openalex.org/A5038235725 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-2451-8598 |
| authorships[1].author.display_name | Frederique Ruf-Zamojski |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I1282927834 |
| authorships[1].affiliations[0].raw_affiliation_string | Cedars-Sinai Medical Center, Department of Medicine, Los Angeles CA |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I98704320 |
| authorships[1].affiliations[1].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[1].institutions[0].id | https://openalex.org/I1282927834 |
| authorships[1].institutions[0].ror | https://ror.org/02pammg90 |
| authorships[1].institutions[0].type | healthcare |
| authorships[1].institutions[0].lineage | https://openalex.org/I1282927834 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Cedars-Sinai Medical Center |
| authorships[1].institutions[1].id | https://openalex.org/I98704320 |
| authorships[1].institutions[1].ror | https://ror.org/04a9tmd77 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[1].institutions[1].country_code | US |
| authorships[1].institutions[1].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Frederique Ruf-zamojski |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Cedars-Sinai Medical Center, Department of Medicine, Los Angeles CA, Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[2].author.id | https://openalex.org/A5081135152 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-7859-3622 |
| authorships[2].author.display_name | Colby T. Ford |
| authorships[2].countries | US |
| authorships[2].affiliations[0].raw_affiliation_string | Tuple LLC, Charlotte, NC, USA |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I102149020 |
| authorships[2].affiliations[1].raw_affiliation_string | University of North Carolina at Charlotte, Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER), Charlotte, NC, USA |
| authorships[2].affiliations[2].institution_ids | https://openalex.org/I102149020 |
| authorships[2].affiliations[2].raw_affiliation_string | University of North Carolina at Charlotte, Department of Bioinformatics and Genomics, Charlotte, NC, USA |
| authorships[2].institutions[0].id | https://openalex.org/I102149020 |
| authorships[2].institutions[0].ror | https://ror.org/04dawnj30 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I102149020 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of North Carolina at Charlotte |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Colby T. Ford |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Tuple LLC, Charlotte, NC, USA, University of North Carolina at Charlotte, Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER), Charlotte, NC, USA, University of North Carolina at Charlotte, Department of Bioinformatics and Genomics, Charlotte, NC, USA |
| authorships[3].author.id | https://openalex.org/A5033246609 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-4165-9990 |
| authorships[3].author.display_name | Pankaj Agarwal |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[3].affiliations[0].raw_affiliation_string | Duke University, Durham, NC |
| authorships[3].institutions[0].id | https://openalex.org/I170897317 |
| authorships[3].institutions[0].ror | https://ror.org/00py81415 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Duke University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Pankaj Agarwal |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Duke University, Durham, NC |
| authorships[4].author.id | https://openalex.org/A5029072664 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-4828-7771 |
| authorships[4].author.display_name | Elena Zaslavsky |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[4].affiliations[0].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[4].institutions[0].id | https://openalex.org/I98704320 |
| authorships[4].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Elena Zaslavsky |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[5].author.id | https://openalex.org/A5073509037 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-5344-5981 |
| authorships[5].author.display_name | German Nudelman |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[5].affiliations[0].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[5].institutions[0].id | https://openalex.org/I98704320 |
| authorships[5].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | German Nudelman |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[6].author.id | https://openalex.org/A5089000748 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-1006-5372 |
| authorships[6].author.display_name | Manoj Hariharan |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1307098950 |
| authorships[6].affiliations[0].raw_affiliation_string | Salk Institute, La Jolla, CA |
| authorships[6].institutions[0].id | https://openalex.org/I1307098950 |
| authorships[6].institutions[0].ror | https://ror.org/03xez1567 |
| authorships[6].institutions[0].type | nonprofit |
| authorships[6].institutions[0].lineage | https://openalex.org/I1307098950 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | Salk Institute for Biological Studies |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Manoj Hariharan |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Salk Institute, La Jolla, CA |
| authorships[7].author.id | https://openalex.org/A5012271592 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | Aliza B. Rubenstein |
| authorships[7].countries | US |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[7].affiliations[0].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[7].institutions[0].id | https://openalex.org/I98704320 |
| authorships[7].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[7].institutions[0].country_code | US |
| authorships[7].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Aliza Rubenstein |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[8].author.id | https://openalex.org/A5062089016 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-4423-3645 |
| authorships[8].author.display_name | Hanna Pinças |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[8].affiliations[0].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[8].institutions[0].id | https://openalex.org/I98704320 |
| authorships[8].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Hanna Pincas |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[9].author.id | https://openalex.org/A5061321094 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-8231-6230 |
| authorships[9].author.display_name | Venugopalan D. Nair |
| authorships[9].countries | US |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[9].affiliations[0].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[9].institutions[0].id | https://openalex.org/I98704320 |
| authorships[9].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[9].institutions[0].country_code | US |
| authorships[9].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Venugopalan D. Nair |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[10].author.id | https://openalex.org/A5016624503 |
| authorships[10].author.orcid | https://orcid.org/0000-0001-7402-8303 |
| authorships[10].author.display_name | Adam Michaleas |
| authorships[10].countries | US |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[10].affiliations[0].raw_affiliation_string | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[10].institutions[0].id | https://openalex.org/I4210122954 |
| authorships[10].institutions[0].ror | https://ror.org/022z6jk58 |
| authorships[10].institutions[0].type | facility |
| authorships[10].institutions[0].lineage | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[10].institutions[0].country_code | US |
| authorships[10].institutions[0].display_name | MIT Lincoln Laboratory |
| authorships[10].institutions[1].id | https://openalex.org/I63966007 |
| authorships[10].institutions[1].ror | https://ror.org/042nb2s44 |
| authorships[10].institutions[1].type | education |
| authorships[10].institutions[1].lineage | https://openalex.org/I63966007 |
| authorships[10].institutions[1].country_code | US |
| authorships[10].institutions[1].display_name | Massachusetts Institute of Technology |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Adam M. Michaleas |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[11].author.id | https://openalex.org/A5058017696 |
| authorships[11].author.orcid | https://orcid.org/0000-0001-5791-1217 |
| authorships[11].author.display_name | Stuart C. Sealfon |
| authorships[11].countries | US |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[11].affiliations[0].raw_affiliation_string | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[11].institutions[0].id | https://openalex.org/I98704320 |
| authorships[11].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[11].institutions[0].type | education |
| authorships[11].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[11].institutions[0].country_code | US |
| authorships[11].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Stuart C. Sealfon |
| authorships[11].is_corresponding | False |
| authorships[11].raw_affiliation_strings | Icahn School of Medicine at Mount Sinai, New York, NY |
| authorships[12].author.id | https://openalex.org/A5007833163 |
| authorships[12].author.orcid | https://orcid.org/0000-0001-7240-2453 |
| authorships[12].author.display_name | Christopher W. Woods |
| authorships[12].countries | US |
| authorships[12].affiliations[0].institution_ids | https://openalex.org/I170897317 |
| authorships[12].affiliations[0].raw_affiliation_string | Duke University, Durham, NC |
| authorships[12].institutions[0].id | https://openalex.org/I170897317 |
| authorships[12].institutions[0].ror | https://ror.org/00py81415 |
| authorships[12].institutions[0].type | education |
| authorships[12].institutions[0].lineage | https://openalex.org/I170897317 |
| authorships[12].institutions[0].country_code | US |
| authorships[12].institutions[0].display_name | Duke University |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Christopher W. Woods |
| authorships[12].is_corresponding | False |
| authorships[12].raw_affiliation_strings | Duke University, Durham, NC |
| authorships[13].author.id | https://openalex.org/A5078675134 |
| authorships[13].author.orcid | https://orcid.org/0000-0001-6479-4899 |
| authorships[13].author.display_name | Kajal Claypool |
| authorships[13].countries | US |
| authorships[13].affiliations[0].institution_ids | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[13].affiliations[0].raw_affiliation_string | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[13].institutions[0].id | https://openalex.org/I4210122954 |
| authorships[13].institutions[0].ror | https://ror.org/022z6jk58 |
| authorships[13].institutions[0].type | facility |
| authorships[13].institutions[0].lineage | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[13].institutions[0].country_code | US |
| authorships[13].institutions[0].display_name | MIT Lincoln Laboratory |
| authorships[13].institutions[1].id | https://openalex.org/I63966007 |
| authorships[13].institutions[1].ror | https://ror.org/042nb2s44 |
| authorships[13].institutions[1].type | education |
| authorships[13].institutions[1].lineage | https://openalex.org/I63966007 |
| authorships[13].institutions[1].country_code | US |
| authorships[13].institutions[1].display_name | Massachusetts Institute of Technology |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Kajal T. Claypool |
| authorships[13].is_corresponding | False |
| authorships[13].raw_affiliation_strings | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[14].author.id | https://openalex.org/A5007734013 |
| authorships[14].author.orcid | https://orcid.org/0000-0002-5493-0399 |
| authorships[14].author.display_name | Rafael Jaimes |
| authorships[14].countries | US |
| authorships[14].affiliations[0].institution_ids | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[14].affiliations[0].raw_affiliation_string | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| authorships[14].institutions[0].id | https://openalex.org/I4210122954 |
| authorships[14].institutions[0].ror | https://ror.org/022z6jk58 |
| authorships[14].institutions[0].type | facility |
| authorships[14].institutions[0].lineage | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| authorships[14].institutions[0].country_code | US |
| authorships[14].institutions[0].display_name | MIT Lincoln Laboratory |
| authorships[14].institutions[1].id | https://openalex.org/I63966007 |
| authorships[14].institutions[1].ror | https://ror.org/042nb2s44 |
| authorships[14].institutions[1].type | education |
| authorships[14].institutions[1].lineage | https://openalex.org/I63966007 |
| authorships[14].institutions[1].country_code | US |
| authorships[14].institutions[1].display_name | Massachusetts Institute of Technology |
| authorships[14].author_position | last |
| authorships[14].raw_author_name | Rafael Jaimes |
| authorships[14].is_corresponding | True |
| authorships[14].raw_affiliation_strings | Lincoln Laboratory, Massachusetts Institute of Technology, Lexington, MA, USA |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Comprehensive Guide of Epigenetics and Transcriptomics Data Quality Control |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T10269 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9991999864578247 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Epigenetics and DNA Methylation |
| related_works | https://openalex.org/W2424092526, https://openalex.org/W2507790420, https://openalex.org/W2212710981, https://openalex.org/W2795223073, https://openalex.org/W4200376839, https://openalex.org/W2151543286, https://openalex.org/W2418239907, https://openalex.org/W2142890267, https://openalex.org/W4280600268, https://openalex.org/W2474086683 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2024.08.02.606411 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2024.08.02.606411 |
| primary_location.id | doi:10.1101/2024.08.02.606411 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2024/08/06/2024.08.02.606411.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2024.08.02.606411 |
| publication_date | 2024-08-06 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2152656267, https://openalex.org/W3088859120, https://openalex.org/W4286824699, https://openalex.org/W2523285199, https://openalex.org/W2017547149, https://openalex.org/W1979594866, https://openalex.org/W2793657715, https://openalex.org/W2949406476, https://openalex.org/W2018604728, https://openalex.org/W3122242878, https://openalex.org/W2117757143, https://openalex.org/W1602702667, https://openalex.org/W1925000049, https://openalex.org/W2206368938, https://openalex.org/W1979674827, https://openalex.org/W2193868205, https://openalex.org/W2162674813, https://openalex.org/W1981509058, https://openalex.org/W2121690903, https://openalex.org/W2407686720, https://openalex.org/W3203959602, https://openalex.org/W4388522770, https://openalex.org/W4210647614, https://openalex.org/W3038923756, https://openalex.org/W2963367647, https://openalex.org/W2236822143, https://openalex.org/W4280528544, https://openalex.org/W2795091386, https://openalex.org/W2002694839, https://openalex.org/W3112677693, https://openalex.org/W2264731258, https://openalex.org/W3157263169, https://openalex.org/W4220941317, https://openalex.org/W3163660840, https://openalex.org/W3102485225, https://openalex.org/W2286142055, https://openalex.org/W3080945506, https://openalex.org/W2883210621, https://openalex.org/W2951506174, https://openalex.org/W1492083415, https://openalex.org/W3201343366, https://openalex.org/W3008816049, https://openalex.org/W2133557974, https://openalex.org/W3036495102, https://openalex.org/W3023623671, https://openalex.org/W2171808845, https://openalex.org/W3126995294, https://openalex.org/W1989338936, https://openalex.org/W2104269895, https://openalex.org/W2114104545, https://openalex.org/W2790768464, https://openalex.org/W2980020683, https://openalex.org/W2572375651, https://openalex.org/W1512733937, https://openalex.org/W2810754857, https://openalex.org/W2889326414, https://openalex.org/W2040149948, https://openalex.org/W4307374479, https://openalex.org/W2529024004, https://openalex.org/W2557182083, https://openalex.org/W1808248494, https://openalex.org/W2971956569, https://openalex.org/W3011994089, https://openalex.org/W2021341670, https://openalex.org/W4291015889, https://openalex.org/W2904347131, https://openalex.org/W2897921044, https://openalex.org/W2152642144, https://openalex.org/W2150127222, https://openalex.org/W2763750537, https://openalex.org/W2127457293 |
| referenced_works_count | 71 |
| abstract_inverted_index.a | 88, 109 |
| abstract_inverted_index.11 | 122 |
| abstract_inverted_index.as | 6 |
| abstract_inverted_index.at | 54 |
| abstract_inverted_index.be | 116 |
| abstract_inverted_index.by | 68 |
| abstract_inverted_index.in | 70 |
| abstract_inverted_index.of | 19, 32, 36, 39, 46, 90, 95, 102, 112, 145 |
| abstract_inverted_index.on | 129 |
| abstract_inverted_index.to | 2, 13, 27, 73, 118, 132, 156 |
| abstract_inverted_index.DNA | 77 |
| abstract_inverted_index.RNA | 75 |
| abstract_inverted_index.The | 142 |
| abstract_inverted_index.and | 8, 16, 30, 42, 82, 125, 136, 149 |
| abstract_inverted_index.are | 140 |
| abstract_inverted_index.can | 10, 22, 115 |
| abstract_inverted_index.for | 153 |
| abstract_inverted_index.has | 65 |
| abstract_inverted_index.the | 14, 28, 37, 40, 59, 103 |
| abstract_inverted_index.Host | 0 |
| abstract_inverted_index.This | 106 |
| abstract_inverted_index.base | 78 |
| abstract_inverted_index.been | 66 |
| abstract_inverted_index.cell | 56 |
| abstract_inverted_index.data | 139 |
| abstract_inverted_index.from | 49, 58, 121 |
| abstract_inverted_index.poor | 137 |
| abstract_inverted_index.same | 60 |
| abstract_inverted_index.such | 5 |
| abstract_inverted_index.that | 98, 114 |
| abstract_inverted_index.used | 117 |
| abstract_inverted_index.with | 25 |
| abstract_inverted_index.agent | 29 |
| abstract_inverted_index.assay | 51 |
| abstract_inverted_index.cause | 11 |
| abstract_inverted_index.guide | 107 |
| abstract_inverted_index.often | 53 |
| abstract_inverted_index.suite | 111 |
| abstract_inverted_index.there | 86 |
| abstract_inverted_index.these | 20 |
| abstract_inverted_index.those | 96 |
| abstract_inverted_index.assay. | 105 |
| abstract_inverted_index.detect | 74 |
| abstract_inverted_index.ensure | 119 |
| abstract_inverted_index.failed | 134 |
| abstract_inverted_index.reveal | 23 |
| abstract_inverted_index.single | 55 |
| abstract_inverted_index.timing | 31 |
| abstract_inverted_index.types, | 52 |
| abstract_inverted_index.address | 133 |
| abstract_inverted_index.assays. | 127 |
| abstract_inverted_index.cascade | 38 |
| abstract_inverted_index.control | 93 |
| abstract_inverted_index.dataset | 147 |
| abstract_inverted_index.enabled | 67 |
| abstract_inverted_index.histone | 80 |
| abstract_inverted_index.metrics | 113, 135 |
| abstract_inverted_index.paucity | 89 |
| abstract_inverted_index.quality | 92, 100, 120, 138, 148 |
| abstract_inverted_index.reflect | 99 |
| abstract_inverted_index.regards | 26 |
| abstract_inverted_index.remains | 87 |
| abstract_inverted_index.results | 155 |
| abstract_inverted_index.Analysis | 18 |
| abstract_inverted_index.However, | 85 |
| abstract_inverted_index.Improved | 62 |
| abstract_inverted_index.accurate | 158 |
| abstract_inverted_index.analysis | 45 |
| abstract_inverted_index.assaying | 71 |
| abstract_inverted_index.consists | 144 |
| abstract_inverted_index.datasets | 48, 97 |
| abstract_inverted_index.exposure | 159 |
| abstract_inverted_index.generate | 157 |
| abstract_inverted_index.improved | 154 |
| abstract_inverted_index.multiple | 50 |
| abstract_inverted_index.outlines | 108 |
| abstract_inverted_index.requires | 44 |
| abstract_inverted_index.response | 1 |
| abstract_inverted_index.rigorous | 91 |
| abstract_inverted_index.workflow | 143 |
| abstract_inverted_index.assessing | 146 |
| abstract_inverted_index.assurance | 101 |
| abstract_inverted_index.benchwork | 151 |
| abstract_inverted_index.chemicals | 9 |
| abstract_inverted_index.chromatin | 83 |
| abstract_inverted_index.different | 123 |
| abstract_inverted_index.discovery | 64 |
| abstract_inverted_index.disparate | 47 |
| abstract_inverted_index.epigenome | 15, 41 |
| abstract_inverted_index.exposure. | 33 |
| abstract_inverted_index.exposures | 4 |
| abstract_inverted_index.pathogens | 7 |
| abstract_inverted_index.protocols | 152 |
| abstract_inverted_index.provided. | 141 |
| abstract_inverted_index.signature | 63 |
| abstract_inverted_index.standards | 94 |
| abstract_inverted_index.Exhaustive | 34 |
| abstract_inverted_index.approaches | 131 |
| abstract_inverted_index.mitigation | 130 |
| abstract_inverted_index.signatures | 24 |
| abstract_inverted_index.techniques | 72 |
| abstract_inverted_index.underlying | 104 |
| abstract_inverted_index.epigenetics | 124 |
| abstract_inverted_index.expression, | 76 |
| abstract_inverted_index.reiterating | 150 |
| abstract_inverted_index.resolution, | 57 |
| abstract_inverted_index.signatures. | 160 |
| abstract_inverted_index.advancements | 69 |
| abstract_inverted_index.biospecimen. | 61 |
| abstract_inverted_index.comprehensive | 110 |
| abstract_inverted_index.environmental | 3 |
| abstract_inverted_index.interrogation | 35 |
| abstract_inverted_index.modifications | 12, 21 |
| abstract_inverted_index.transcriptome | 43 |
| abstract_inverted_index.accessibility. | 84 |
| abstract_inverted_index.modifications, | 79, 81 |
| abstract_inverted_index.transcriptome. | 17 |
| abstract_inverted_index.Recommendations | 128 |
| abstract_inverted_index.transcriptomics | 126 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5007734013 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 15 |
| corresponding_institution_ids | https://openalex.org/I4210122954, https://openalex.org/I63966007 |
| citation_normalized_percentile.value | 0.14188399 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |