Creating cell-specific computational models of stem cell-derived cardiomyocytes using optical experiments Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pcbi.1011806
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pcbi.1011806
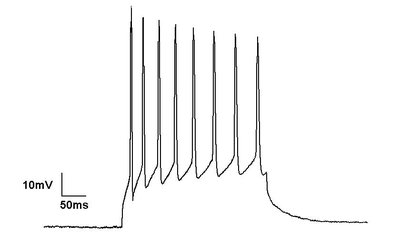
Human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) have gained traction as a powerful model in cardiac disease and therapeutics research, since iPSCs are self-renewing and can be derived from healthy and diseased patients without invasive surgery. However, current iPSC-CM differentiation methods produce cardiomyocytes with immature, fetal-like electrophysiological phenotypes, and the variety of maturation protocols in the literature results in phenotypic differences between labs. Heterogeneity of iPSC donor genetic backgrounds contributes to additional phenotypic variability. Several mathematical models of iPSC-CM electrophysiology have been developed to help to predict cell responses, but these models individually do not capture the phenotypic variability observed in iPSC-CMs. Here, we tackle these limitations by developing a computational pipeline to calibrate cell preparation-specific iPSC-CM electrophysiological parameters. We used the genetic algorithm (GA), a heuristic parameter calibration method, to tune ion channel parameters in a mathematical model of iPSC-CM physiology. To systematically optimize an experimental protocol that generates sufficient data for parameter calibration, we created in silico datasets by simulating various protocols applied to a population of models with known conductance variations, and then fitted parameters to those datasets. We found that calibrating to voltage and calcium transient data under 3 varied experimental conditions, including electrical pacing combined with ion channel blockade and changing buffer ion concentrations, improved model parameter estimates and model predictions of unseen channel block responses. This observation also held when the fitted data were normalized, suggesting that normalized fluorescence recordings, which are more accessible and higher throughput than patch clamp recordings, could sufficiently inform conductance parameters. Therefore, this computational pipeline can be applied to different iPSC-CM preparations to determine cell line-specific ion channel properties and understand the mechanisms behind variability in perturbation responses.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.pcbi.1011806
- OA Status
- gold
- Cited By
- 5
- References
- 62
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4402437249
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4402437249Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.pcbi.1011806Digital Object Identifier
- Title
-
Creating cell-specific computational models of stem cell-derived cardiomyocytes using optical experimentsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-09-11Full publication date if available
- Authors
-
Janice Yang, Neil Daily, Taylor K. Pullinger, Tetsuro Wakatsuki, Eric A. SobieList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.pcbi.1011806Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1371/journal.pcbi.1011806Direct OA link when available
- Concepts
-
Induced pluripotent stem cell, In silico, Cardiac electrophysiology, Electrophysiology, Biological system, Population, Computational biology, Computer science, Biology, Computational model, Genetic model, Bioinformatics, Neuroscience, Artificial intelligence, Embryonic stem cell, Genetics, Medicine, Environmental health, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
5Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 2Per-year citation counts (last 5 years)
- References (count)
-
62Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4402437249 |
|---|---|
| doi | https://doi.org/10.1371/journal.pcbi.1011806 |
| ids.doi | https://doi.org/10.1371/journal.pcbi.1011806 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39259757 |
| ids.openalex | https://openalex.org/W4402437249 |
| fwci | 2.63564357 |
| mesh[0].qualifier_ui | Q000502 |
| mesh[0].descriptor_ui | D032383 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | physiology |
| mesh[0].descriptor_name | Myocytes, Cardiac |
| mesh[1].qualifier_ui | Q000166 |
| mesh[1].descriptor_ui | D032383 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | cytology |
| mesh[1].descriptor_name | Myocytes, Cardiac |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D006801 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Humans |
| mesh[3].qualifier_ui | Q000166 |
| mesh[3].descriptor_ui | D057026 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | cytology |
| mesh[3].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[4].qualifier_ui | Q000502 |
| mesh[4].descriptor_ui | D057026 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | physiology |
| mesh[4].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[5].qualifier_ui | Q000502 |
| mesh[5].descriptor_ui | D002454 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | physiology |
| mesh[5].descriptor_name | Cell Differentiation |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D003198 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Computer Simulation |
| mesh[7].qualifier_ui | Q000379 |
| mesh[7].descriptor_ui | D019295 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | methods |
| mesh[7].descriptor_name | Computational Biology |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D000465 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Algorithms |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D007473 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Ion Channels |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D008955 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Models, Cardiovascular |
| mesh[11].qualifier_ui | Q000502 |
| mesh[11].descriptor_ui | D032383 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | physiology |
| mesh[11].descriptor_name | Myocytes, Cardiac |
| mesh[12].qualifier_ui | Q000166 |
| mesh[12].descriptor_ui | D032383 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | cytology |
| mesh[12].descriptor_name | Myocytes, Cardiac |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D006801 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Humans |
| mesh[14].qualifier_ui | Q000166 |
| mesh[14].descriptor_ui | D057026 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | cytology |
| mesh[14].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[15].qualifier_ui | Q000502 |
| mesh[15].descriptor_ui | D057026 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | physiology |
| mesh[15].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[16].qualifier_ui | Q000502 |
| mesh[16].descriptor_ui | D002454 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | physiology |
| mesh[16].descriptor_name | Cell Differentiation |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D003198 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Computer Simulation |
| mesh[18].qualifier_ui | Q000379 |
| mesh[18].descriptor_ui | D019295 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | methods |
| mesh[18].descriptor_name | Computational Biology |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D000465 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Algorithms |
| mesh[20].qualifier_ui | Q000378 |
| mesh[20].descriptor_ui | D007473 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | metabolism |
| mesh[20].descriptor_name | Ion Channels |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D008955 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Models, Cardiovascular |
| mesh[22].qualifier_ui | Q000502 |
| mesh[22].descriptor_ui | D032383 |
| mesh[22].is_major_topic | True |
| mesh[22].qualifier_name | physiology |
| mesh[22].descriptor_name | Myocytes, Cardiac |
| mesh[23].qualifier_ui | Q000166 |
| mesh[23].descriptor_ui | D032383 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | cytology |
| mesh[23].descriptor_name | Myocytes, Cardiac |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D006801 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Humans |
| mesh[25].qualifier_ui | Q000166 |
| mesh[25].descriptor_ui | D057026 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | cytology |
| mesh[25].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[26].qualifier_ui | Q000502 |
| mesh[26].descriptor_ui | D057026 |
| mesh[26].is_major_topic | True |
| mesh[26].qualifier_name | physiology |
| mesh[26].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[27].qualifier_ui | Q000502 |
| mesh[27].descriptor_ui | D002454 |
| mesh[27].is_major_topic | True |
| mesh[27].qualifier_name | physiology |
| mesh[27].descriptor_name | Cell Differentiation |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D003198 |
| mesh[28].is_major_topic | True |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Computer Simulation |
| mesh[29].qualifier_ui | Q000379 |
| mesh[29].descriptor_ui | D019295 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | methods |
| mesh[29].descriptor_name | Computational Biology |
| mesh[30].qualifier_ui | |
| mesh[30].descriptor_ui | D000465 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | |
| mesh[30].descriptor_name | Algorithms |
| mesh[31].qualifier_ui | Q000378 |
| mesh[31].descriptor_ui | D007473 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | metabolism |
| mesh[31].descriptor_name | Ion Channels |
| mesh[32].qualifier_ui | |
| mesh[32].descriptor_ui | D008955 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | |
| mesh[32].descriptor_name | Models, Cardiovascular |
| mesh[33].qualifier_ui | Q000502 |
| mesh[33].descriptor_ui | D032383 |
| mesh[33].is_major_topic | True |
| mesh[33].qualifier_name | physiology |
| mesh[33].descriptor_name | Myocytes, Cardiac |
| mesh[34].qualifier_ui | Q000166 |
| mesh[34].descriptor_ui | D032383 |
| mesh[34].is_major_topic | True |
| mesh[34].qualifier_name | cytology |
| mesh[34].descriptor_name | Myocytes, Cardiac |
| mesh[35].qualifier_ui | |
| mesh[35].descriptor_ui | D006801 |
| mesh[35].is_major_topic | False |
| mesh[35].qualifier_name | |
| mesh[35].descriptor_name | Humans |
| mesh[36].qualifier_ui | Q000166 |
| mesh[36].descriptor_ui | D057026 |
| mesh[36].is_major_topic | True |
| mesh[36].qualifier_name | cytology |
| mesh[36].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[37].qualifier_ui | Q000502 |
| mesh[37].descriptor_ui | D057026 |
| mesh[37].is_major_topic | True |
| mesh[37].qualifier_name | physiology |
| mesh[37].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[38].qualifier_ui | Q000502 |
| mesh[38].descriptor_ui | D002454 |
| mesh[38].is_major_topic | True |
| mesh[38].qualifier_name | physiology |
| mesh[38].descriptor_name | Cell Differentiation |
| mesh[39].qualifier_ui | |
| mesh[39].descriptor_ui | D003198 |
| mesh[39].is_major_topic | True |
| mesh[39].qualifier_name | |
| mesh[39].descriptor_name | Computer Simulation |
| mesh[40].qualifier_ui | Q000379 |
| mesh[40].descriptor_ui | D019295 |
| mesh[40].is_major_topic | False |
| mesh[40].qualifier_name | methods |
| mesh[40].descriptor_name | Computational Biology |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D000465 |
| mesh[41].is_major_topic | False |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | Algorithms |
| mesh[42].qualifier_ui | Q000378 |
| mesh[42].descriptor_ui | D007473 |
| mesh[42].is_major_topic | False |
| mesh[42].qualifier_name | metabolism |
| mesh[42].descriptor_name | Ion Channels |
| mesh[43].qualifier_ui | |
| mesh[43].descriptor_ui | D008955 |
| mesh[43].is_major_topic | False |
| mesh[43].qualifier_name | |
| mesh[43].descriptor_name | Models, Cardiovascular |
| mesh[44].qualifier_ui | Q000502 |
| mesh[44].descriptor_ui | D032383 |
| mesh[44].is_major_topic | True |
| mesh[44].qualifier_name | physiology |
| mesh[44].descriptor_name | Myocytes, Cardiac |
| mesh[45].qualifier_ui | Q000166 |
| mesh[45].descriptor_ui | D032383 |
| mesh[45].is_major_topic | True |
| mesh[45].qualifier_name | cytology |
| mesh[45].descriptor_name | Myocytes, Cardiac |
| mesh[46].qualifier_ui | |
| mesh[46].descriptor_ui | D006801 |
| mesh[46].is_major_topic | False |
| mesh[46].qualifier_name | |
| mesh[46].descriptor_name | Humans |
| mesh[47].qualifier_ui | Q000166 |
| mesh[47].descriptor_ui | D057026 |
| mesh[47].is_major_topic | True |
| mesh[47].qualifier_name | cytology |
| mesh[47].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[48].qualifier_ui | Q000502 |
| mesh[48].descriptor_ui | D057026 |
| mesh[48].is_major_topic | True |
| mesh[48].qualifier_name | physiology |
| mesh[48].descriptor_name | Induced Pluripotent Stem Cells |
| mesh[49].qualifier_ui | Q000502 |
| mesh[49].descriptor_ui | D002454 |
| mesh[49].is_major_topic | True |
| mesh[49].qualifier_name | physiology |
| mesh[49].descriptor_name | Cell Differentiation |
| type | article |
| title | Creating cell-specific computational models of stem cell-derived cardiomyocytes using optical experiments |
| awards[0].id | https://openalex.org/G5730666829 |
| awards[0].funder_id | https://openalex.org/F4320337472 |
| awards[0].display_name | |
| awards[0].funder_award_id | ULTR004419 |
| awards[0].funder_display_name | National Center for Advancing Translational Sciences |
| awards[1].id | https://openalex.org/G3062774485 |
| awards[1].funder_id | https://openalex.org/F4320337611 |
| awards[1].display_name | |
| awards[1].funder_award_id | T32 HD 075735 |
| awards[1].funder_display_name | Eunice Kennedy Shriver National Institute of Child Health and Human Development |
| awards[2].id | https://openalex.org/G7738174925 |
| awards[2].funder_id | https://openalex.org/F4320337338 |
| awards[2].display_name | |
| awards[2].funder_award_id | R44 HL 139248 |
| awards[2].funder_display_name | National Heart, Lung, and Blood Institute |
| biblio.issue | 9 |
| biblio.volume | 20 |
| biblio.last_page | e1011806 |
| biblio.first_page | e1011806 |
| topics[0].id | https://openalex.org/T11601 |
| topics[0].field.id | https://openalex.org/fields/28 |
| topics[0].field.display_name | Neuroscience |
| topics[0].score | 0.9980999827384949 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2804 |
| topics[0].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[0].display_name | Neuroscience and Neural Engineering |
| topics[1].id | https://openalex.org/T10505 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9937999844551086 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Pluripotent Stem Cells Research |
| topics[2].id | https://openalex.org/T10217 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9918000102043152 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2705 |
| topics[2].subfield.display_name | Cardiology and Cardiovascular Medicine |
| topics[2].display_name | Cardiac electrophysiology and arrhythmias |
| funders[0].id | https://openalex.org/F4320337338 |
| funders[0].ror | https://ror.org/012pb6c26 |
| funders[0].display_name | National Heart, Lung, and Blood Institute |
| funders[1].id | https://openalex.org/F4320337472 |
| funders[1].ror | https://ror.org/04pw6fb54 |
| funders[1].display_name | National Center for Advancing Translational Sciences |
| funders[2].id | https://openalex.org/F4320337611 |
| funders[2].ror | https://ror.org/04byxyr05 |
| funders[2].display_name | Eunice Kennedy Shriver National Institute of Child Health and Human Development |
| is_xpac | False |
| apc_list.value | 2655 |
| apc_list.currency | USD |
| apc_list.value_usd | 2655 |
| apc_paid.value | 2655 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2655 |
| concepts[0].id | https://openalex.org/C107459253 |
| concepts[0].level | 4 |
| concepts[0].score | 0.7045862674713135 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q674925 |
| concepts[0].display_name | Induced pluripotent stem cell |
| concepts[1].id | https://openalex.org/C2775905019 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6275668144226074 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q192572 |
| concepts[1].display_name | In silico |
| concepts[2].id | https://openalex.org/C61381695 |
| concepts[2].level | 3 |
| concepts[2].score | 0.591711699962616 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q3302130 |
| concepts[2].display_name | Cardiac electrophysiology |
| concepts[3].id | https://openalex.org/C185263204 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5123361349105835 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1154774 |
| concepts[3].display_name | Electrophysiology |
| concepts[4].id | https://openalex.org/C186060115 |
| concepts[4].level | 1 |
| concepts[4].score | 0.4983348846435547 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q30336093 |
| concepts[4].display_name | Biological system |
| concepts[5].id | https://openalex.org/C2908647359 |
| concepts[5].level | 2 |
| concepts[5].score | 0.4865800142288208 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q2625603 |
| concepts[5].display_name | Population |
| concepts[6].id | https://openalex.org/C70721500 |
| concepts[6].level | 1 |
| concepts[6].score | 0.48584267497062683 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[6].display_name | Computational biology |
| concepts[7].id | https://openalex.org/C41008148 |
| concepts[7].level | 0 |
| concepts[7].score | 0.4646799862384796 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[7].display_name | Computer science |
| concepts[8].id | https://openalex.org/C86803240 |
| concepts[8].level | 0 |
| concepts[8].score | 0.448074609041214 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[8].display_name | Biology |
| concepts[9].id | https://openalex.org/C66024118 |
| concepts[9].level | 2 |
| concepts[9].score | 0.42963477969169617 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1122506 |
| concepts[9].display_name | Computational model |
| concepts[10].id | https://openalex.org/C2992519594 |
| concepts[10].level | 3 |
| concepts[10].score | 0.4151984751224518 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q870433 |
| concepts[10].display_name | Genetic model |
| concepts[11].id | https://openalex.org/C60644358 |
| concepts[11].level | 1 |
| concepts[11].score | 0.3582616448402405 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[11].display_name | Bioinformatics |
| concepts[12].id | https://openalex.org/C169760540 |
| concepts[12].level | 1 |
| concepts[12].score | 0.26389920711517334 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q207011 |
| concepts[12].display_name | Neuroscience |
| concepts[13].id | https://openalex.org/C154945302 |
| concepts[13].level | 1 |
| concepts[13].score | 0.22441497445106506 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[13].display_name | Artificial intelligence |
| concepts[14].id | https://openalex.org/C145103041 |
| concepts[14].level | 3 |
| concepts[14].score | 0.20670974254608154 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q1151519 |
| concepts[14].display_name | Embryonic stem cell |
| concepts[15].id | https://openalex.org/C54355233 |
| concepts[15].level | 1 |
| concepts[15].score | 0.1333204209804535 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[15].display_name | Genetics |
| concepts[16].id | https://openalex.org/C71924100 |
| concepts[16].level | 0 |
| concepts[16].score | 0.10556107759475708 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[16].display_name | Medicine |
| concepts[17].id | https://openalex.org/C99454951 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q932068 |
| concepts[17].display_name | Environmental health |
| concepts[18].id | https://openalex.org/C104317684 |
| concepts[18].level | 2 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[18].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/induced-pluripotent-stem-cell |
| keywords[0].score | 0.7045862674713135 |
| keywords[0].display_name | Induced pluripotent stem cell |
| keywords[1].id | https://openalex.org/keywords/in-silico |
| keywords[1].score | 0.6275668144226074 |
| keywords[1].display_name | In silico |
| keywords[2].id | https://openalex.org/keywords/cardiac-electrophysiology |
| keywords[2].score | 0.591711699962616 |
| keywords[2].display_name | Cardiac electrophysiology |
| keywords[3].id | https://openalex.org/keywords/electrophysiology |
| keywords[3].score | 0.5123361349105835 |
| keywords[3].display_name | Electrophysiology |
| keywords[4].id | https://openalex.org/keywords/biological-system |
| keywords[4].score | 0.4983348846435547 |
| keywords[4].display_name | Biological system |
| keywords[5].id | https://openalex.org/keywords/population |
| keywords[5].score | 0.4865800142288208 |
| keywords[5].display_name | Population |
| keywords[6].id | https://openalex.org/keywords/computational-biology |
| keywords[6].score | 0.48584267497062683 |
| keywords[6].display_name | Computational biology |
| keywords[7].id | https://openalex.org/keywords/computer-science |
| keywords[7].score | 0.4646799862384796 |
| keywords[7].display_name | Computer science |
| keywords[8].id | https://openalex.org/keywords/biology |
| keywords[8].score | 0.448074609041214 |
| keywords[8].display_name | Biology |
| keywords[9].id | https://openalex.org/keywords/computational-model |
| keywords[9].score | 0.42963477969169617 |
| keywords[9].display_name | Computational model |
| keywords[10].id | https://openalex.org/keywords/genetic-model |
| keywords[10].score | 0.4151984751224518 |
| keywords[10].display_name | Genetic model |
| keywords[11].id | https://openalex.org/keywords/bioinformatics |
| keywords[11].score | 0.3582616448402405 |
| keywords[11].display_name | Bioinformatics |
| keywords[12].id | https://openalex.org/keywords/neuroscience |
| keywords[12].score | 0.26389920711517334 |
| keywords[12].display_name | Neuroscience |
| keywords[13].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[13].score | 0.22441497445106506 |
| keywords[13].display_name | Artificial intelligence |
| keywords[14].id | https://openalex.org/keywords/embryonic-stem-cell |
| keywords[14].score | 0.20670974254608154 |
| keywords[14].display_name | Embryonic stem cell |
| keywords[15].id | https://openalex.org/keywords/genetics |
| keywords[15].score | 0.1333204209804535 |
| keywords[15].display_name | Genetics |
| keywords[16].id | https://openalex.org/keywords/medicine |
| keywords[16].score | 0.10556107759475708 |
| keywords[16].display_name | Medicine |
| language | en |
| locations[0].id | doi:10.1371/journal.pcbi.1011806 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S86033158 |
| locations[0].source.issn | 1553-734X, 1553-7358 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1553-734X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS Computational Biology |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_lineage_names | Public Library of Science |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS Computational Biology |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.pcbi.1011806 |
| locations[1].id | pmid:39259757 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PLoS computational biology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39259757 |
| locations[2].id | pmh:oai:RePEc:plo:pcbi00:1011806 |
| locations[2].is_oa | False |
| locations[2].source.id | https://openalex.org/S4306401271 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | RePEc: Research Papers in Economics |
| locations[2].source.host_organization | https://openalex.org/I77793887 |
| locations[2].source.host_organization_name | Federal Reserve Bank of St. Louis |
| locations[2].source.host_organization_lineage | https://openalex.org/I77793887 |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1011806 |
| locations[3].id | pmh:oai:doaj.org/article:0e07087b5ff44ebaaf53e12cb8627162 |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401280 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | PLoS Computational Biology, Vol 20, Iss 9, p e1011806 (2024) |
| locations[3].landing_page_url | https://doaj.org/article/0e07087b5ff44ebaaf53e12cb8627162 |
| locations[4].id | pmh:oai:pubmedcentral.nih.gov:11460686 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S2764455111 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | PubMed Central |
| locations[4].source.host_organization | https://openalex.org/I1299303238 |
| locations[4].source.host_organization_name | National Institutes of Health |
| locations[4].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[4].license | other-oa |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/other-oa |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | PLoS Comput Biol |
| locations[4].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11460686 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5111108006 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Janice Yang |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Pharmacological Sciences & Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America. |
| authorships[0].institutions[0].id | https://openalex.org/I98704320 |
| authorships[0].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Janice Yang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Pharmacological Sciences & Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America. |
| authorships[1].author.id | https://openalex.org/A5074703289 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-9494-6603 |
| authorships[1].author.display_name | Neil Daily |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210166000 |
| authorships[1].affiliations[0].raw_affiliation_string | InvivoSciences Inc., Madison, Wisconsin, United States of America. |
| authorships[1].institutions[0].id | https://openalex.org/I4210166000 |
| authorships[1].institutions[0].ror | https://ror.org/05wfxya04 |
| authorships[1].institutions[0].type | company |
| authorships[1].institutions[0].lineage | https://openalex.org/I4210166000 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Invivo Sciences (United States) |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Neil J Daily |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | InvivoSciences Inc., Madison, Wisconsin, United States of America. |
| authorships[2].author.id | https://openalex.org/A5040571558 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-4821-8047 |
| authorships[2].author.display_name | Taylor K. Pullinger |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Pharmacological Sciences & Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America. |
| authorships[2].institutions[0].id | https://openalex.org/I98704320 |
| authorships[2].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Taylor K Pullinger |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Pharmacological Sciences & Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America. |
| authorships[3].author.id | https://openalex.org/A5039100833 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-2175-1132 |
| authorships[3].author.display_name | Tetsuro Wakatsuki |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I4210166000 |
| authorships[3].affiliations[0].raw_affiliation_string | InvivoSciences Inc., Madison, Wisconsin, United States of America. |
| authorships[3].institutions[0].id | https://openalex.org/I4210166000 |
| authorships[3].institutions[0].ror | https://ror.org/05wfxya04 |
| authorships[3].institutions[0].type | company |
| authorships[3].institutions[0].lineage | https://openalex.org/I4210166000 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Invivo Sciences (United States) |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Tetsuro Wakatsuki |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | InvivoSciences Inc., Madison, Wisconsin, United States of America. |
| authorships[4].author.id | https://openalex.org/A5000485841 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-7327-8538 |
| authorships[4].author.display_name | Eric A. Sobie |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I98704320 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Pharmacological Sciences & Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America. |
| authorships[4].institutions[0].id | https://openalex.org/I98704320 |
| authorships[4].institutions[0].ror | https://ror.org/04a9tmd77 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I1320796813, https://openalex.org/I98704320 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Icahn School of Medicine at Mount Sinai |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Eric A Sobie |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Pharmacological Sciences & Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1371/journal.pcbi.1011806 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Creating cell-specific computational models of stem cell-derived cardiomyocytes using optical experiments |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-23T05:10:03.516525 |
| primary_topic.id | https://openalex.org/T11601 |
| primary_topic.field.id | https://openalex.org/fields/28 |
| primary_topic.field.display_name | Neuroscience |
| primary_topic.score | 0.9980999827384949 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2804 |
| primary_topic.subfield.display_name | Cellular and Molecular Neuroscience |
| primary_topic.display_name | Neuroscience and Neural Engineering |
| related_works | https://openalex.org/W3129816939, https://openalex.org/W2912862995, https://openalex.org/W2985427592, https://openalex.org/W2101501829, https://openalex.org/W2400569965, https://openalex.org/W4392388912, https://openalex.org/W2071953960, https://openalex.org/W2262354220, https://openalex.org/W2087732651, https://openalex.org/W1988916448 |
| cited_by_count | 5 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1371/journal.pcbi.1011806 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S86033158 |
| best_oa_location.source.issn | 1553-734X, 1553-7358 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1553-734X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS Computational Biology |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_lineage_names | Public Library of Science |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS Computational Biology |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.pcbi.1011806 |
| primary_location.id | doi:10.1371/journal.pcbi.1011806 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S86033158 |
| primary_location.source.issn | 1553-734X, 1553-7358 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1553-734X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS Computational Biology |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_lineage_names | Public Library of Science |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS Computational Biology |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.pcbi.1011806 |
| publication_date | 2024-09-11 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W1907511822, https://openalex.org/W2982460272, https://openalex.org/W3137052139, https://openalex.org/W4200395889, https://openalex.org/W2041761134, https://openalex.org/W3011609945, https://openalex.org/W3004299218, https://openalex.org/W3138368030, https://openalex.org/W3015790912, https://openalex.org/W3093465023, https://openalex.org/W3130249007, https://openalex.org/W2033765226, https://openalex.org/W2622073064, https://openalex.org/W3198981533, https://openalex.org/W3188617807, https://openalex.org/W1974119618, https://openalex.org/W2786635794, https://openalex.org/W2803772876, https://openalex.org/W2773865883, https://openalex.org/W2956154393, https://openalex.org/W1974946304, https://openalex.org/W2808838901, https://openalex.org/W3132825319, https://openalex.org/W2044253779, https://openalex.org/W2156639802, https://openalex.org/W2114156487, https://openalex.org/W2538021067, https://openalex.org/W3172672014, https://openalex.org/W2949360571, https://openalex.org/W2106871016, https://openalex.org/W4389436538, https://openalex.org/W3014696819, https://openalex.org/W4210813151, https://openalex.org/W2071953960, https://openalex.org/W2141260882, https://openalex.org/W2138728269, https://openalex.org/W1965139977, https://openalex.org/W2279387088, https://openalex.org/W2170990019, https://openalex.org/W2487405781, https://openalex.org/W2779105341, https://openalex.org/W4283789469, https://openalex.org/W2092692908, https://openalex.org/W2014018052, https://openalex.org/W2101484286, https://openalex.org/W3007670977, https://openalex.org/W2963113143, https://openalex.org/W2075402097, https://openalex.org/W2746858283, https://openalex.org/W2952336269, https://openalex.org/W3212410380, https://openalex.org/W2092950144, https://openalex.org/W3113024012, https://openalex.org/W2161304688, https://openalex.org/W2105437542, https://openalex.org/W2086215140, https://openalex.org/W3137121888, https://openalex.org/W2895910955, https://openalex.org/W2166305181, https://openalex.org/W2890760570, https://openalex.org/W4205919873, https://openalex.org/W2951315938 |
| referenced_works_count | 62 |
| abstract_inverted_index.3 | 192 |
| abstract_inverted_index.a | 11, 109, 125, 136, 166 |
| abstract_inverted_index.To | 142 |
| abstract_inverted_index.We | 119, 181 |
| abstract_inverted_index.an | 145 |
| abstract_inverted_index.as | 10 |
| abstract_inverted_index.be | 26, 257 |
| abstract_inverted_index.by | 107, 160 |
| abstract_inverted_index.do | 93 |
| abstract_inverted_index.in | 14, 54, 58, 100, 135, 157, 276 |
| abstract_inverted_index.of | 51, 64, 77, 139, 168, 216 |
| abstract_inverted_index.to | 70, 83, 85, 112, 130, 165, 178, 185, 259, 263 |
| abstract_inverted_index.we | 103, 155 |
| abstract_inverted_index.and | 17, 24, 30, 48, 174, 187, 204, 213, 240, 270 |
| abstract_inverted_index.are | 22, 237 |
| abstract_inverted_index.but | 89 |
| abstract_inverted_index.can | 25, 256 |
| abstract_inverted_index.for | 152 |
| abstract_inverted_index.ion | 132, 201, 207, 267 |
| abstract_inverted_index.not | 94 |
| abstract_inverted_index.the | 49, 55, 96, 121, 226, 272 |
| abstract_inverted_index.This | 221 |
| abstract_inverted_index.also | 223 |
| abstract_inverted_index.been | 81 |
| abstract_inverted_index.cell | 87, 114, 265 |
| abstract_inverted_index.data | 151, 190, 228 |
| abstract_inverted_index.from | 28 |
| abstract_inverted_index.have | 7, 80 |
| abstract_inverted_index.held | 224 |
| abstract_inverted_index.help | 84 |
| abstract_inverted_index.iPSC | 65 |
| abstract_inverted_index.more | 238 |
| abstract_inverted_index.stem | 3 |
| abstract_inverted_index.than | 243 |
| abstract_inverted_index.that | 148, 183, 232 |
| abstract_inverted_index.then | 175 |
| abstract_inverted_index.this | 253 |
| abstract_inverted_index.tune | 131 |
| abstract_inverted_index.used | 120 |
| abstract_inverted_index.were | 229 |
| abstract_inverted_index.when | 225 |
| abstract_inverted_index.with | 43, 170, 200 |
| abstract_inverted_index.(GA), | 124 |
| abstract_inverted_index.Here, | 102 |
| abstract_inverted_index.Human | 0 |
| abstract_inverted_index.block | 219 |
| abstract_inverted_index.clamp | 245 |
| abstract_inverted_index.could | 247 |
| abstract_inverted_index.donor | 66 |
| abstract_inverted_index.found | 182 |
| abstract_inverted_index.iPSCs | 21 |
| abstract_inverted_index.known | 171 |
| abstract_inverted_index.labs. | 62 |
| abstract_inverted_index.model | 13, 138, 210, 214 |
| abstract_inverted_index.patch | 244 |
| abstract_inverted_index.since | 20 |
| abstract_inverted_index.these | 90, 105 |
| abstract_inverted_index.those | 179 |
| abstract_inverted_index.under | 191 |
| abstract_inverted_index.which | 236 |
| abstract_inverted_index.behind | 274 |
| abstract_inverted_index.buffer | 206 |
| abstract_inverted_index.fitted | 176, 227 |
| abstract_inverted_index.gained | 8 |
| abstract_inverted_index.higher | 241 |
| abstract_inverted_index.inform | 249 |
| abstract_inverted_index.models | 76, 91, 169 |
| abstract_inverted_index.pacing | 198 |
| abstract_inverted_index.silico | 158 |
| abstract_inverted_index.tackle | 104 |
| abstract_inverted_index.unseen | 217 |
| abstract_inverted_index.varied | 193 |
| abstract_inverted_index.Several | 74 |
| abstract_inverted_index.applied | 164, 258 |
| abstract_inverted_index.between | 61 |
| abstract_inverted_index.calcium | 188 |
| abstract_inverted_index.capture | 95 |
| abstract_inverted_index.cardiac | 15 |
| abstract_inverted_index.channel | 133, 202, 218, 268 |
| abstract_inverted_index.created | 156 |
| abstract_inverted_index.current | 37 |
| abstract_inverted_index.derived | 27 |
| abstract_inverted_index.disease | 16 |
| abstract_inverted_index.genetic | 67, 122 |
| abstract_inverted_index.healthy | 29 |
| abstract_inverted_index.iPSC-CM | 38, 78, 116, 140, 261 |
| abstract_inverted_index.induced | 1 |
| abstract_inverted_index.method, | 129 |
| abstract_inverted_index.methods | 40 |
| abstract_inverted_index.predict | 86 |
| abstract_inverted_index.produce | 41 |
| abstract_inverted_index.results | 57 |
| abstract_inverted_index.variety | 50 |
| abstract_inverted_index.various | 162 |
| abstract_inverted_index.voltage | 186 |
| abstract_inverted_index.without | 33 |
| abstract_inverted_index.However, | 36 |
| abstract_inverted_index.blockade | 203 |
| abstract_inverted_index.changing | 205 |
| abstract_inverted_index.combined | 199 |
| abstract_inverted_index.datasets | 159 |
| abstract_inverted_index.diseased | 31 |
| abstract_inverted_index.improved | 209 |
| abstract_inverted_index.invasive | 34 |
| abstract_inverted_index.observed | 99 |
| abstract_inverted_index.optimize | 144 |
| abstract_inverted_index.patients | 32 |
| abstract_inverted_index.pipeline | 111, 255 |
| abstract_inverted_index.powerful | 12 |
| abstract_inverted_index.protocol | 147 |
| abstract_inverted_index.surgery. | 35 |
| abstract_inverted_index.traction | 9 |
| abstract_inverted_index.algorithm | 123 |
| abstract_inverted_index.calibrate | 113 |
| abstract_inverted_index.datasets. | 180 |
| abstract_inverted_index.determine | 264 |
| abstract_inverted_index.developed | 82 |
| abstract_inverted_index.different | 260 |
| abstract_inverted_index.estimates | 212 |
| abstract_inverted_index.generates | 149 |
| abstract_inverted_index.heuristic | 126 |
| abstract_inverted_index.iPSC-CMs. | 101 |
| abstract_inverted_index.immature, | 44 |
| abstract_inverted_index.including | 196 |
| abstract_inverted_index.parameter | 127, 153, 211 |
| abstract_inverted_index.protocols | 53, 163 |
| abstract_inverted_index.research, | 19 |
| abstract_inverted_index.transient | 189 |
| abstract_inverted_index.(iPSC-CMs) | 6 |
| abstract_inverted_index.Therefore, | 252 |
| abstract_inverted_index.accessible | 239 |
| abstract_inverted_index.additional | 71 |
| abstract_inverted_index.developing | 108 |
| abstract_inverted_index.electrical | 197 |
| abstract_inverted_index.fetal-like | 45 |
| abstract_inverted_index.literature | 56 |
| abstract_inverted_index.maturation | 52 |
| abstract_inverted_index.mechanisms | 273 |
| abstract_inverted_index.normalized | 233 |
| abstract_inverted_index.parameters | 134, 177 |
| abstract_inverted_index.phenotypic | 59, 72, 97 |
| abstract_inverted_index.population | 167 |
| abstract_inverted_index.properties | 269 |
| abstract_inverted_index.responses, | 88 |
| abstract_inverted_index.responses. | 220, 278 |
| abstract_inverted_index.simulating | 161 |
| abstract_inverted_index.sufficient | 150 |
| abstract_inverted_index.suggesting | 231 |
| abstract_inverted_index.throughput | 242 |
| abstract_inverted_index.understand | 271 |
| abstract_inverted_index.backgrounds | 68 |
| abstract_inverted_index.calibrating | 184 |
| abstract_inverted_index.calibration | 128 |
| abstract_inverted_index.conditions, | 195 |
| abstract_inverted_index.conductance | 172, 250 |
| abstract_inverted_index.contributes | 69 |
| abstract_inverted_index.differences | 60 |
| abstract_inverted_index.limitations | 106 |
| abstract_inverted_index.normalized, | 230 |
| abstract_inverted_index.observation | 222 |
| abstract_inverted_index.parameters. | 118, 251 |
| abstract_inverted_index.phenotypes, | 47 |
| abstract_inverted_index.physiology. | 141 |
| abstract_inverted_index.pluripotent | 2 |
| abstract_inverted_index.predictions | 215 |
| abstract_inverted_index.recordings, | 235, 246 |
| abstract_inverted_index.variability | 98, 275 |
| abstract_inverted_index.variations, | 173 |
| abstract_inverted_index.calibration, | 154 |
| abstract_inverted_index.cell-derived | 4 |
| abstract_inverted_index.experimental | 146, 194 |
| abstract_inverted_index.fluorescence | 234 |
| abstract_inverted_index.individually | 92 |
| abstract_inverted_index.mathematical | 75, 137 |
| abstract_inverted_index.perturbation | 277 |
| abstract_inverted_index.preparations | 262 |
| abstract_inverted_index.sufficiently | 248 |
| abstract_inverted_index.therapeutics | 18 |
| abstract_inverted_index.variability. | 73 |
| abstract_inverted_index.Heterogeneity | 63 |
| abstract_inverted_index.computational | 110, 254 |
| abstract_inverted_index.line-specific | 266 |
| abstract_inverted_index.self-renewing | 23 |
| abstract_inverted_index.cardiomyocytes | 5, 42 |
| abstract_inverted_index.systematically | 143 |
| abstract_inverted_index.concentrations, | 208 |
| abstract_inverted_index.differentiation | 39 |
| abstract_inverted_index.electrophysiology | 79 |
| abstract_inverted_index.electrophysiological | 46, 117 |
| abstract_inverted_index.preparation-specific | 115 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 94 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| citation_normalized_percentile.value | 0.85122074 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |