Current status of antimicrobial peptides databases and computational tools for optimization Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.36953/ecj.29252932
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.36953/ecj.29252932
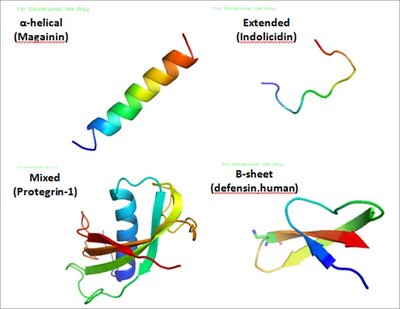
Antimicrobial resistance is projected as next pandemic with a propensity of around 10 million deaths per year by 2050. Alternatives of the antibiotic compounds are required to be explored on priority basis to treat viral, bacterial, and fungal infections. Antimicrobial peptides (AMPs) are emerging as a promising therapeutic alternative to combat serious microbial infections. Plenty of natural AMPs have been isolated and characterized from different sources posing a need of categorizing them in various databases. Bioinspired from natural peptides, several synthetic AMPs have been designed and tested against microbial pathogens. An ideal antimicrobial peptide needs to have specific physical and structural properties. To analyse structure, biological activity and toxicity of the AMPs, different computational tools have been developed which are available in the public domain accelerating the research and development of antimicrobial peptides. This review encompasses properties, structure and databases of antimicrobial peptide with a key focus on computational tools designed for prediction of structure, function and toxicity of the synthetic and natural peptides.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.36953/ecj.29252932
- OA Status
- gold
- Cited By
- 1
- References
- 88
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4408666566
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4408666566Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.36953/ecj.29252932Digital Object Identifier
- Title
-
Current status of antimicrobial peptides databases and computational tools for optimizationWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-02-05Full publication date if available
- Authors
-
Madhulika Jha, Akash Nautiyal, Kumud Pant, Navin KumarList of authors in order
- Landing page
-
https://doi.org/10.36953/ecj.29252932Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.36953/ecj.29252932Direct OA link when available
- Concepts
-
Antimicrobial peptides, Antimicrobial, Computer science, Current (fluid), Database, Data science, Computational biology, Biology, Microbiology, Engineering, Electrical engineeringTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
88Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4408666566 |
|---|---|
| doi | https://doi.org/10.36953/ecj.29252932 |
| ids.doi | https://doi.org/10.36953/ecj.29252932 |
| ids.openalex | https://openalex.org/W4408666566 |
| fwci | 3.54425088 |
| type | article |
| title | Current status of antimicrobial peptides databases and computational tools for optimization |
| biblio.issue | 1 |
| biblio.volume | 26 |
| biblio.last_page | 292 |
| biblio.first_page | 281 |
| topics[0].id | https://openalex.org/T11103 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2404 |
| topics[0].subfield.display_name | Microbiology |
| topics[0].display_name | Antimicrobial Peptides and Activities |
| topics[1].id | https://openalex.org/T13326 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9988999962806702 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Biochemical and Structural Characterization |
| topics[2].id | https://openalex.org/T12576 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9909999966621399 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | vaccines and immunoinformatics approaches |
| is_xpac | False |
| apc_list.value | 7500 |
| apc_list.currency | INR |
| apc_list.value_usd | 225 |
| apc_paid.value | 7500 |
| apc_paid.currency | INR |
| apc_paid.value_usd | 225 |
| concepts[0].id | https://openalex.org/C540938839 |
| concepts[0].level | 3 |
| concepts[0].score | 0.532012939453125 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1201508 |
| concepts[0].display_name | Antimicrobial peptides |
| concepts[1].id | https://openalex.org/C4937899 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5104626417160034 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q68541106 |
| concepts[1].display_name | Antimicrobial |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.4714321196079254 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C148043351 |
| concepts[3].level | 2 |
| concepts[3].score | 0.4274721145629883 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q4456944 |
| concepts[3].display_name | Current (fluid) |
| concepts[4].id | https://openalex.org/C77088390 |
| concepts[4].level | 1 |
| concepts[4].score | 0.4272553026676178 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q8513 |
| concepts[4].display_name | Database |
| concepts[5].id | https://openalex.org/C2522767166 |
| concepts[5].level | 1 |
| concepts[5].score | 0.4055745005607605 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q2374463 |
| concepts[5].display_name | Data science |
| concepts[6].id | https://openalex.org/C70721500 |
| concepts[6].level | 1 |
| concepts[6].score | 0.36047500371932983 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[6].display_name | Computational biology |
| concepts[7].id | https://openalex.org/C86803240 |
| concepts[7].level | 0 |
| concepts[7].score | 0.2825871706008911 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[7].display_name | Biology |
| concepts[8].id | https://openalex.org/C89423630 |
| concepts[8].level | 1 |
| concepts[8].score | 0.19325321912765503 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[8].display_name | Microbiology |
| concepts[9].id | https://openalex.org/C127413603 |
| concepts[9].level | 0 |
| concepts[9].score | 0.16180479526519775 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q11023 |
| concepts[9].display_name | Engineering |
| concepts[10].id | https://openalex.org/C119599485 |
| concepts[10].level | 1 |
| concepts[10].score | 0.0 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q43035 |
| concepts[10].display_name | Electrical engineering |
| keywords[0].id | https://openalex.org/keywords/antimicrobial-peptides |
| keywords[0].score | 0.532012939453125 |
| keywords[0].display_name | Antimicrobial peptides |
| keywords[1].id | https://openalex.org/keywords/antimicrobial |
| keywords[1].score | 0.5104626417160034 |
| keywords[1].display_name | Antimicrobial |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.4714321196079254 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/current |
| keywords[3].score | 0.4274721145629883 |
| keywords[3].display_name | Current (fluid) |
| keywords[4].id | https://openalex.org/keywords/database |
| keywords[4].score | 0.4272553026676178 |
| keywords[4].display_name | Database |
| keywords[5].id | https://openalex.org/keywords/data-science |
| keywords[5].score | 0.4055745005607605 |
| keywords[5].display_name | Data science |
| keywords[6].id | https://openalex.org/keywords/computational-biology |
| keywords[6].score | 0.36047500371932983 |
| keywords[6].display_name | Computational biology |
| keywords[7].id | https://openalex.org/keywords/biology |
| keywords[7].score | 0.2825871706008911 |
| keywords[7].display_name | Biology |
| keywords[8].id | https://openalex.org/keywords/microbiology |
| keywords[8].score | 0.19325321912765503 |
| keywords[8].display_name | Microbiology |
| keywords[9].id | https://openalex.org/keywords/engineering |
| keywords[9].score | 0.16180479526519775 |
| keywords[9].display_name | Engineering |
| language | en |
| locations[0].id | doi:10.36953/ecj.29252932 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2764520314 |
| locations[0].source.issn | 0972-3099, 2278-5124 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 0972-3099 |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Environment Conservation Journal |
| locations[0].source.host_organization | |
| locations[0].source.host_organization_name | |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Environment Conservation Journal |
| locations[0].landing_page_url | https://doi.org/10.36953/ecj.29252932 |
| locations[1].id | pmh:oai:doaj.org/article:3ce2de75da6d40ef9e1fc6aae0e2166d |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306401280 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | article |
| locations[1].license_id | |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | Environment Conservation Journal, Vol 26, Iss 1 (2025) |
| locations[1].landing_page_url | https://doaj.org/article/3ce2de75da6d40ef9e1fc6aae0e2166d |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5066429434 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Madhulika Jha |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Madhulika Jha |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5116720040 |
| authorships[1].author.orcid | https://orcid.org/0009-0001-1130-7679 |
| authorships[1].author.display_name | Akash Nautiyal |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Akash Nautiyal |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5073511309 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-0193-6604 |
| authorships[2].author.display_name | Kumud Pant |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Kumud Pant |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5057237463 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-3531-7414 |
| authorships[3].author.display_name | Navin Kumar |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Navin Kumar |
| authorships[3].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.36953/ecj.29252932 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Current status of antimicrobial peptides databases and computational tools for optimization |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11103 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2404 |
| primary_topic.subfield.display_name | Microbiology |
| primary_topic.display_name | Antimicrobial Peptides and Activities |
| related_works | https://openalex.org/W2358368915, https://openalex.org/W2380741856, https://openalex.org/W2348658803, https://openalex.org/W1973192249, https://openalex.org/W2354039930, https://openalex.org/W4375855538, https://openalex.org/W3087020980, https://openalex.org/W2107525095, https://openalex.org/W4211241221, https://openalex.org/W2511368942 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 2 |
| best_oa_location.id | doi:10.36953/ecj.29252932 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2764520314 |
| best_oa_location.source.issn | 0972-3099, 2278-5124 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 0972-3099 |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Environment Conservation Journal |
| best_oa_location.source.host_organization | |
| best_oa_location.source.host_organization_name | |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Environment Conservation Journal |
| best_oa_location.landing_page_url | https://doi.org/10.36953/ecj.29252932 |
| primary_location.id | doi:10.36953/ecj.29252932 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2764520314 |
| primary_location.source.issn | 0972-3099, 2278-5124 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 0972-3099 |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Environment Conservation Journal |
| primary_location.source.host_organization | |
| primary_location.source.host_organization_name | |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Environment Conservation Journal |
| primary_location.landing_page_url | https://doi.org/10.36953/ecj.29252932 |
| publication_date | 2025-02-05 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W2792378492, https://openalex.org/W4210968711, https://openalex.org/W2789307963, https://openalex.org/W3097657320, https://openalex.org/W2102567112, https://openalex.org/W2896223271, https://openalex.org/W3207784306, https://openalex.org/W3133998099, https://openalex.org/W4386473409, https://openalex.org/W4287377229, https://openalex.org/W2039155298, https://openalex.org/W3010686554, https://openalex.org/W4319296069, https://openalex.org/W2128495797, https://openalex.org/W2097099115, https://openalex.org/W1992450378, https://openalex.org/W2617456826, https://openalex.org/W4308834846, https://openalex.org/W2076860935, https://openalex.org/W2553564063, https://openalex.org/W4316041410, https://openalex.org/W2551912621, https://openalex.org/W1994025057, https://openalex.org/W3138220359, https://openalex.org/W4283585440, https://openalex.org/W2746050631, https://openalex.org/W2124846900, https://openalex.org/W3092909579, https://openalex.org/W2064283997, https://openalex.org/W4286493084, https://openalex.org/W2899153547, https://openalex.org/W3216979156, https://openalex.org/W1978444624, https://openalex.org/W2965128681, https://openalex.org/W2936682441, https://openalex.org/W2083416546, https://openalex.org/W3094708881, https://openalex.org/W2067402644, https://openalex.org/W4362520112, https://openalex.org/W1986388841, https://openalex.org/W4280599277, https://openalex.org/W2303974462, https://openalex.org/W2563632637, https://openalex.org/W237973401, https://openalex.org/W1999679502, https://openalex.org/W2146289378, https://openalex.org/W4210250306, https://openalex.org/W1980150325, https://openalex.org/W2588467130, https://openalex.org/W2947485682, https://openalex.org/W2557546772, https://openalex.org/W4212915338, https://openalex.org/W4294292553, https://openalex.org/W2035832348, https://openalex.org/W2084845164, https://openalex.org/W2727386932, https://openalex.org/W2130873227, https://openalex.org/W2152380569, https://openalex.org/W3097405028, https://openalex.org/W2080104470, https://openalex.org/W2945010589, https://openalex.org/W2065618112, https://openalex.org/W2903234151, https://openalex.org/W2098368706, https://openalex.org/W1863757753, https://openalex.org/W2129615006, https://openalex.org/W3188372451, https://openalex.org/W3200790559, https://openalex.org/W3155913164, https://openalex.org/W2033873633, https://openalex.org/W2009031433, https://openalex.org/W2114247676, https://openalex.org/W4212867833, https://openalex.org/W2980919412, https://openalex.org/W3017661193, https://openalex.org/W2163316049, https://openalex.org/W2175270296, https://openalex.org/W2778198694, https://openalex.org/W1982551127, https://openalex.org/W1989731705, https://openalex.org/W1998064689, https://openalex.org/W4308370073, https://openalex.org/W4207019670, https://openalex.org/W3080137602, https://openalex.org/W2023758688, https://openalex.org/W2015490529, https://openalex.org/W4226455819, https://openalex.org/W4225733893 |
| referenced_works_count | 88 |
| abstract_inverted_index.a | 8, 45, 67, 144 |
| abstract_inverted_index.10 | 12 |
| abstract_inverted_index.An | 90 |
| abstract_inverted_index.To | 102 |
| abstract_inverted_index.as | 4, 44 |
| abstract_inverted_index.be | 27 |
| abstract_inverted_index.by | 17 |
| abstract_inverted_index.in | 72, 121 |
| abstract_inverted_index.is | 2 |
| abstract_inverted_index.of | 10, 20, 55, 69, 109, 130, 140, 153, 158 |
| abstract_inverted_index.on | 29, 147 |
| abstract_inverted_index.to | 26, 32, 49, 95 |
| abstract_inverted_index.and | 36, 61, 85, 99, 107, 128, 138, 156, 161 |
| abstract_inverted_index.are | 24, 42, 119 |
| abstract_inverted_index.for | 151 |
| abstract_inverted_index.key | 145 |
| abstract_inverted_index.per | 15 |
| abstract_inverted_index.the | 21, 110, 122, 126, 159 |
| abstract_inverted_index.AMPs | 57, 81 |
| abstract_inverted_index.This | 133 |
| abstract_inverted_index.been | 59, 83, 116 |
| abstract_inverted_index.from | 63, 76 |
| abstract_inverted_index.have | 58, 82, 96, 115 |
| abstract_inverted_index.need | 68 |
| abstract_inverted_index.next | 5 |
| abstract_inverted_index.them | 71 |
| abstract_inverted_index.with | 7, 143 |
| abstract_inverted_index.year | 16 |
| abstract_inverted_index.2050. | 18 |
| abstract_inverted_index.AMPs, | 111 |
| abstract_inverted_index.basis | 31 |
| abstract_inverted_index.focus | 146 |
| abstract_inverted_index.ideal | 91 |
| abstract_inverted_index.needs | 94 |
| abstract_inverted_index.tools | 114, 149 |
| abstract_inverted_index.treat | 33 |
| abstract_inverted_index.which | 118 |
| abstract_inverted_index.(AMPs) | 41 |
| abstract_inverted_index.Plenty | 54 |
| abstract_inverted_index.around | 11 |
| abstract_inverted_index.combat | 50 |
| abstract_inverted_index.deaths | 14 |
| abstract_inverted_index.domain | 124 |
| abstract_inverted_index.fungal | 37 |
| abstract_inverted_index.posing | 66 |
| abstract_inverted_index.public | 123 |
| abstract_inverted_index.review | 134 |
| abstract_inverted_index.tested | 86 |
| abstract_inverted_index.viral, | 34 |
| abstract_inverted_index.against | 87 |
| abstract_inverted_index.analyse | 103 |
| abstract_inverted_index.million | 13 |
| abstract_inverted_index.natural | 56, 77, 162 |
| abstract_inverted_index.peptide | 93, 142 |
| abstract_inverted_index.serious | 51 |
| abstract_inverted_index.several | 79 |
| abstract_inverted_index.sources | 65 |
| abstract_inverted_index.various | 73 |
| abstract_inverted_index.activity | 106 |
| abstract_inverted_index.designed | 84, 150 |
| abstract_inverted_index.emerging | 43 |
| abstract_inverted_index.explored | 28 |
| abstract_inverted_index.function | 155 |
| abstract_inverted_index.isolated | 60 |
| abstract_inverted_index.pandemic | 6 |
| abstract_inverted_index.peptides | 40 |
| abstract_inverted_index.physical | 98 |
| abstract_inverted_index.priority | 30 |
| abstract_inverted_index.required | 25 |
| abstract_inverted_index.research | 127 |
| abstract_inverted_index.specific | 97 |
| abstract_inverted_index.toxicity | 108, 157 |
| abstract_inverted_index.available | 120 |
| abstract_inverted_index.compounds | 23 |
| abstract_inverted_index.databases | 139 |
| abstract_inverted_index.developed | 117 |
| abstract_inverted_index.different | 64, 112 |
| abstract_inverted_index.microbial | 52, 88 |
| abstract_inverted_index.peptides, | 78 |
| abstract_inverted_index.peptides. | 132, 163 |
| abstract_inverted_index.projected | 3 |
| abstract_inverted_index.promising | 46 |
| abstract_inverted_index.structure | 137 |
| abstract_inverted_index.synthetic | 80, 160 |
| abstract_inverted_index.antibiotic | 22 |
| abstract_inverted_index.bacterial, | 35 |
| abstract_inverted_index.biological | 105 |
| abstract_inverted_index.databases. | 74 |
| abstract_inverted_index.pathogens. | 89 |
| abstract_inverted_index.prediction | 152 |
| abstract_inverted_index.propensity | 9 |
| abstract_inverted_index.resistance | 1 |
| abstract_inverted_index.structural | 100 |
| abstract_inverted_index.structure, | 104, 154 |
| abstract_inverted_index.Bioinspired | 75 |
| abstract_inverted_index.alternative | 48 |
| abstract_inverted_index.development | 129 |
| abstract_inverted_index.encompasses | 135 |
| abstract_inverted_index.infections. | 38, 53 |
| abstract_inverted_index.properties, | 136 |
| abstract_inverted_index.properties. | 101 |
| abstract_inverted_index.therapeutic | 47 |
| abstract_inverted_index.Alternatives | 19 |
| abstract_inverted_index.accelerating | 125 |
| abstract_inverted_index.categorizing | 70 |
| abstract_inverted_index.Antimicrobial | 0, 39 |
| abstract_inverted_index.antimicrobial | 92, 131, 141 |
| abstract_inverted_index.characterized | 62 |
| abstract_inverted_index.computational | 113, 148 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 4 |
| citation_normalized_percentile.value | 0.82350906 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |