Data from Dual targeting of apoptotic and signaling pathways in T-lineage acute lymphoblastic leukemia Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1158/1078-0432.c.6740540.v5
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1158/1078-0432.c.6740540.v5
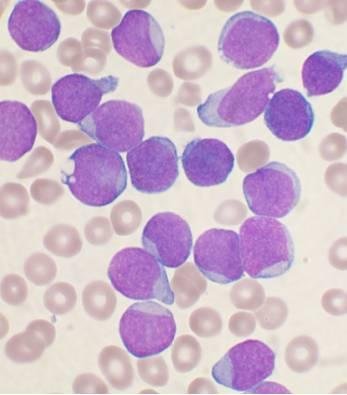
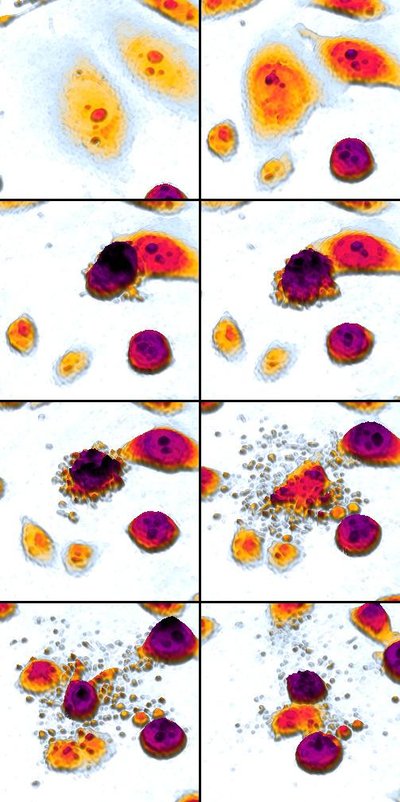
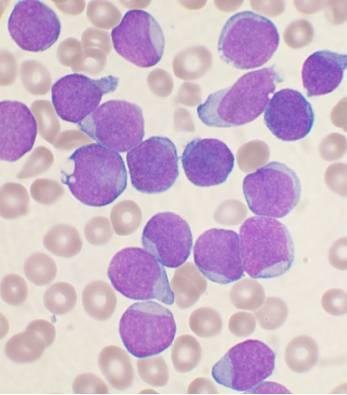
Purpose: Relapsed T-acute lymphoblastic leukemia (T-ALL) has limited treatment options. We investigated mechanisms of resistance to BH3 mimetics in T-ALL in order to develop rational combination strategies. We also looked at the preclinical efficacy of NWP-0476, a novel BCL-2/BCL-xL inhibitor, as single agent and combination therapy in T-ALL. Methods: We used BH3 profiling as a predictive tool for BH3 mimetic response in T-ALL. Using isogenic control, venetoclax-resistant (ven-R) and NWP-0476-resistant (NWP-R) cells, phosphokinase array was performed to identify differentially regulated signaling pathways. Results: Typical T-ALL cells had increased dependence on BCL-xL, while early T-precursor (ETP)-ALL cells had higher BCL-2 dependence for survival. BCL-2/BCL-xL dual inhibitors were effective against both subtypes of T-lineage ALL. A 71-protein human phosphokinase array showed increased LCK activity in ven-R cells, and increased ACK1 activity in ven-R and NWP-R cells. We hypothesized that pre-TCR and ACK1 signaling pathways are drivers of resistance to BCL-2 and BCL-xL inhibition, respectively. First, we silenced LCK gene in T-ALL cell lines, which resulted in increased sensitivity to BCL-2 inhibition. Mechanistically, LCK activated NF-κB pathway and the expression of BCL-xL. Silencing ACK1 gene resulted in increased sensitivity to both BCL-2 and BCL-xL inhibitors. ACK1 signaling up-regulated AKT pathway, which inhibited the pro-apoptotic function of BAD. In a T-ALL patient-derived xenograft model, combination of NWP-0476 and dasatinib demonstrated synergy without major organ toxicity. Conclusions:LCK and ACK1 signaling pathways are critical regulators of BH3 mimetic resistance in T-ALL. Combination of BH3 mimetics with tyrosine kinase inhibitors might be effective against relapsed T-ALL.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1158/1078-0432.c.6740540.v5
- OA Status
- gold
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4402818171
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4402818171Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1158/1078-0432.c.6740540.v5Digital Object Identifier
- Title
-
Data from Dual targeting of apoptotic and signaling pathways in T-lineage acute lymphoblastic leukemiaWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-09-16Full publication date if available
- Authors
-
Caner Saygin, Giorgia Giordano, Kathryn Shimamoto, Bart Eisfelder, Anika T. Thomas-Toth, Girish Venkataraman, Vijayalakshmi Ananthanarayanan, Tiffaney L. Vincent, Adam DuVall, Anand Patel, Yi Chen, Fenlai Tan, Stephen P. Anthony, Yu Chen, Yue Shen, Olatoyosi Odenike, David T. Teachey, Barbara L. Kee, James L. LaBelle, Wendy StockList of authors in order
- Landing page
-
https://doi.org/10.1158/1078-0432.c.6740540.v5Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1158/1078-0432.c.6740540.v5Direct OA link when available
- Concepts
-
Lymphoblastic Leukemia, Lineage (genetic), Cancer research, Apoptosis, Dual (grammatical number), Signal transduction, Leukemia, Medicine, Biology, Immunology, Cell biology, Genetics, Gene, Literature, ArtTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4402818171 |
|---|---|
| doi | https://doi.org/10.1158/1078-0432.c.6740540.v5 |
| ids.doi | https://doi.org/10.1158/1078-0432.c.6740540.v5 |
| ids.openalex | https://openalex.org/W4402818171 |
| fwci | 0.0 |
| type | preprint |
| title | Data from Dual targeting of apoptotic and signaling pathways in T-lineage acute lymphoblastic leukemia |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10950 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9478999972343445 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2739 |
| topics[0].subfield.display_name | Public Health, Environmental and Occupational Health |
| topics[0].display_name | Acute Lymphoblastic Leukemia research |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2909962599 |
| concepts[0].level | 3 |
| concepts[0].score | 0.790176510810852 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q180664 |
| concepts[0].display_name | Lymphoblastic Leukemia |
| concepts[1].id | https://openalex.org/C2776817793 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6913534998893738 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q6553369 |
| concepts[1].display_name | Lineage (genetic) |
| concepts[2].id | https://openalex.org/C502942594 |
| concepts[2].level | 1 |
| concepts[2].score | 0.6223227977752686 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q3421914 |
| concepts[2].display_name | Cancer research |
| concepts[3].id | https://openalex.org/C190283241 |
| concepts[3].level | 2 |
| concepts[3].score | 0.6122555732727051 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q14599311 |
| concepts[3].display_name | Apoptosis |
| concepts[4].id | https://openalex.org/C2780980858 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5151852369308472 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q110022 |
| concepts[4].display_name | Dual (grammatical number) |
| concepts[5].id | https://openalex.org/C62478195 |
| concepts[5].level | 2 |
| concepts[5].score | 0.49794769287109375 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q828130 |
| concepts[5].display_name | Signal transduction |
| concepts[6].id | https://openalex.org/C2778461978 |
| concepts[6].level | 2 |
| concepts[6].score | 0.38473448157310486 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q29496 |
| concepts[6].display_name | Leukemia |
| concepts[7].id | https://openalex.org/C71924100 |
| concepts[7].level | 0 |
| concepts[7].score | 0.36681830883026123 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[7].display_name | Medicine |
| concepts[8].id | https://openalex.org/C86803240 |
| concepts[8].level | 0 |
| concepts[8].score | 0.34671151638031006 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[8].display_name | Biology |
| concepts[9].id | https://openalex.org/C203014093 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3199867308139801 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q101929 |
| concepts[9].display_name | Immunology |
| concepts[10].id | https://openalex.org/C95444343 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3096621036529541 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[10].display_name | Cell biology |
| concepts[11].id | https://openalex.org/C54355233 |
| concepts[11].level | 1 |
| concepts[11].score | 0.192834734916687 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[11].display_name | Genetics |
| concepts[12].id | https://openalex.org/C104317684 |
| concepts[12].level | 2 |
| concepts[12].score | 0.152163565158844 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[12].display_name | Gene |
| concepts[13].id | https://openalex.org/C124952713 |
| concepts[13].level | 1 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q8242 |
| concepts[13].display_name | Literature |
| concepts[14].id | https://openalex.org/C142362112 |
| concepts[14].level | 0 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q735 |
| concepts[14].display_name | Art |
| keywords[0].id | https://openalex.org/keywords/lymphoblastic-leukemia |
| keywords[0].score | 0.790176510810852 |
| keywords[0].display_name | Lymphoblastic Leukemia |
| keywords[1].id | https://openalex.org/keywords/lineage |
| keywords[1].score | 0.6913534998893738 |
| keywords[1].display_name | Lineage (genetic) |
| keywords[2].id | https://openalex.org/keywords/cancer-research |
| keywords[2].score | 0.6223227977752686 |
| keywords[2].display_name | Cancer research |
| keywords[3].id | https://openalex.org/keywords/apoptosis |
| keywords[3].score | 0.6122555732727051 |
| keywords[3].display_name | Apoptosis |
| keywords[4].id | https://openalex.org/keywords/dual |
| keywords[4].score | 0.5151852369308472 |
| keywords[4].display_name | Dual (grammatical number) |
| keywords[5].id | https://openalex.org/keywords/signal-transduction |
| keywords[5].score | 0.49794769287109375 |
| keywords[5].display_name | Signal transduction |
| keywords[6].id | https://openalex.org/keywords/leukemia |
| keywords[6].score | 0.38473448157310486 |
| keywords[6].display_name | Leukemia |
| keywords[7].id | https://openalex.org/keywords/medicine |
| keywords[7].score | 0.36681830883026123 |
| keywords[7].display_name | Medicine |
| keywords[8].id | https://openalex.org/keywords/biology |
| keywords[8].score | 0.34671151638031006 |
| keywords[8].display_name | Biology |
| keywords[9].id | https://openalex.org/keywords/immunology |
| keywords[9].score | 0.3199867308139801 |
| keywords[9].display_name | Immunology |
| keywords[10].id | https://openalex.org/keywords/cell-biology |
| keywords[10].score | 0.3096621036529541 |
| keywords[10].display_name | Cell biology |
| keywords[11].id | https://openalex.org/keywords/genetics |
| keywords[11].score | 0.192834734916687 |
| keywords[11].display_name | Genetics |
| keywords[12].id | https://openalex.org/keywords/gene |
| keywords[12].score | 0.152163565158844 |
| keywords[12].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1158/1078-0432.c.6740540.v5 |
| locations[0].is_oa | True |
| locations[0].source | |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1158/1078-0432.c.6740540.v5 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5003321703 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-7617-8356 |
| authorships[0].author.display_name | Caner Saygin |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Caner Saygin |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5082721654 |
| authorships[1].author.orcid | https://orcid.org/0009-0004-7473-5883 |
| authorships[1].author.display_name | Giorgia Giordano |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Giorgia Giordano |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5046147043 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-2732-6495 |
| authorships[2].author.display_name | Kathryn Shimamoto |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Kathryn Shimamoto |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5109622698 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Bart Eisfelder |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Bart Eisfelder |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5030982107 |
| authorships[4].author.orcid | https://orcid.org/0009-0007-4231-8227 |
| authorships[4].author.display_name | Anika T. Thomas-Toth |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Anika Thomas-Toth |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5042629306 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-8674-2608 |
| authorships[5].author.display_name | Girish Venkataraman |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Girish Venkataraman |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5031283803 |
| authorships[6].author.orcid | https://orcid.org/0009-0001-3993-4404 |
| authorships[6].author.display_name | Vijayalakshmi Ananthanarayanan |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Vijayalakshmi Ananthanarayanan |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5076506220 |
| authorships[7].author.orcid | https://orcid.org/0009-0004-6147-7088 |
| authorships[7].author.display_name | Tiffaney L. Vincent |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Tiffaney L. Vincent |
| authorships[7].is_corresponding | False |
| authorships[8].author.id | https://openalex.org/A5058055734 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-9163-8792 |
| authorships[8].author.display_name | Adam DuVall |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Adam DuVall |
| authorships[8].is_corresponding | False |
| authorships[9].author.id | https://openalex.org/A5038290613 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-0296-8686 |
| authorships[9].author.display_name | Anand Patel |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Anand A. Patel |
| authorships[9].is_corresponding | False |
| authorships[10].author.id | https://openalex.org/A5092272362 |
| authorships[10].author.orcid | https://orcid.org/0009-0005-0945-6930 |
| authorships[10].author.display_name | Yi Chen |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Yi Chen |
| authorships[10].is_corresponding | False |
| authorships[11].author.id | https://openalex.org/A5032952436 |
| authorships[11].author.orcid | https://orcid.org/0009-0002-7289-3218 |
| authorships[11].author.display_name | Fenlai Tan |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Fenlai Tan |
| authorships[11].is_corresponding | False |
| authorships[12].author.id | https://openalex.org/A5029811877 |
| authorships[12].author.orcid | https://orcid.org/0009-0009-6005-5592 |
| authorships[12].author.display_name | Stephen P. Anthony |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Stephen P. Anthony |
| authorships[12].is_corresponding | False |
| authorships[13].author.id | https://openalex.org/A5100694803 |
| authorships[13].author.orcid | https://orcid.org/0009-0000-5165-2912 |
| authorships[13].author.display_name | Yu Chen |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Yu Chen |
| authorships[13].is_corresponding | False |
| authorships[14].author.id | https://openalex.org/A5030176308 |
| authorships[14].author.orcid | https://orcid.org/0009-0004-8991-7612 |
| authorships[14].author.display_name | Yue Shen |
| authorships[14].author_position | middle |
| authorships[14].raw_author_name | Yue Shen |
| authorships[14].is_corresponding | False |
| authorships[15].author.id | https://openalex.org/A5060015628 |
| authorships[15].author.orcid | https://orcid.org/0000-0002-4027-4124 |
| authorships[15].author.display_name | Olatoyosi Odenike |
| authorships[15].author_position | middle |
| authorships[15].raw_author_name | Olatoyosi Odenike |
| authorships[15].is_corresponding | False |
| authorships[16].author.id | https://openalex.org/A5028647628 |
| authorships[16].author.orcid | https://orcid.org/0000-0001-7373-8987 |
| authorships[16].author.display_name | David T. Teachey |
| authorships[16].author_position | middle |
| authorships[16].raw_author_name | David T. Teachey |
| authorships[16].is_corresponding | False |
| authorships[17].author.id | https://openalex.org/A5088061651 |
| authorships[17].author.orcid | https://orcid.org/0000-0001-7014-1982 |
| authorships[17].author.display_name | Barbara L. Kee |
| authorships[17].author_position | middle |
| authorships[17].raw_author_name | Barbara L. Kee |
| authorships[17].is_corresponding | False |
| authorships[18].author.id | https://openalex.org/A5035243936 |
| authorships[18].author.orcid | https://orcid.org/0000-0001-6776-4695 |
| authorships[18].author.display_name | James L. LaBelle |
| authorships[18].author_position | middle |
| authorships[18].raw_author_name | James LaBelle |
| authorships[18].is_corresponding | False |
| authorships[19].author.id | https://openalex.org/A5005939966 |
| authorships[19].author.orcid | https://orcid.org/0000-0002-8349-9200 |
| authorships[19].author.display_name | Wendy Stock |
| authorships[19].author_position | last |
| authorships[19].raw_author_name | Wendy Stock |
| authorships[19].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1158/1078-0432.c.6740540.v5 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Data from Dual targeting of apoptotic and signaling pathways in T-lineage acute lymphoblastic leukemia |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10950 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9478999972343445 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2739 |
| primary_topic.subfield.display_name | Public Health, Environmental and Occupational Health |
| primary_topic.display_name | Acute Lymphoblastic Leukemia research |
| related_works | https://openalex.org/W2506149209, https://openalex.org/W2352153050, https://openalex.org/W2140071436, https://openalex.org/W2393965794, https://openalex.org/W2091678252, https://openalex.org/W4246442859, https://openalex.org/W4298086599, https://openalex.org/W4246926659, https://openalex.org/W1972233104, https://openalex.org/W2321211866 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1158/1078-0432.c.6740540.v5 |
| best_oa_location.is_oa | True |
| best_oa_location.source | |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1158/1078-0432.c.6740540.v5 |
| primary_location.id | doi:10.1158/1078-0432.c.6740540.v5 |
| primary_location.is_oa | True |
| primary_location.source | |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1158/1078-0432.c.6740540.v5 |
| publication_date | 2024-09-16 |
| publication_year | 2024 |
| referenced_works_count | 0 |
| abstract_inverted_index.A | 113 |
| abstract_inverted_index.a | 36, 54, 205 |
| abstract_inverted_index.In | 204 |
| abstract_inverted_index.We | 10, 27, 49, 134 |
| abstract_inverted_index.as | 40, 53 |
| abstract_inverted_index.at | 30 |
| abstract_inverted_index.be | 244 |
| abstract_inverted_index.in | 18, 20, 46, 61, 122, 129, 157, 163, 183, 233 |
| abstract_inverted_index.of | 13, 34, 110, 144, 177, 202, 211, 229, 236 |
| abstract_inverted_index.on | 89 |
| abstract_inverted_index.to | 15, 22, 76, 146, 166, 186 |
| abstract_inverted_index.we | 153 |
| abstract_inverted_index.AKT | 195 |
| abstract_inverted_index.BH3 | 16, 51, 58, 230, 237 |
| abstract_inverted_index.LCK | 120, 155, 170 |
| abstract_inverted_index.and | 43, 68, 125, 131, 138, 148, 174, 189, 213, 222 |
| abstract_inverted_index.are | 142, 226 |
| abstract_inverted_index.for | 57, 100 |
| abstract_inverted_index.had | 86, 96 |
| abstract_inverted_index.has | 6 |
| abstract_inverted_index.the | 31, 175, 199 |
| abstract_inverted_index.was | 74 |
| abstract_inverted_index.ACK1 | 127, 139, 180, 192, 223 |
| abstract_inverted_index.ALL. | 112 |
| abstract_inverted_index.BAD. | 203 |
| abstract_inverted_index.also | 28 |
| abstract_inverted_index.both | 108, 187 |
| abstract_inverted_index.cell | 159 |
| abstract_inverted_index.dual | 103 |
| abstract_inverted_index.gene | 156, 181 |
| abstract_inverted_index.that | 136 |
| abstract_inverted_index.tool | 56 |
| abstract_inverted_index.used | 50 |
| abstract_inverted_index.were | 105 |
| abstract_inverted_index.with | 239 |
| abstract_inverted_index.BCL-2 | 98, 147, 167, 188 |
| abstract_inverted_index.NWP-R | 132 |
| abstract_inverted_index.T-ALL | 19, 84, 158, 206 |
| abstract_inverted_index.Using | 63 |
| abstract_inverted_index.agent | 42 |
| abstract_inverted_index.array | 73, 117 |
| abstract_inverted_index.cells | 85, 95 |
| abstract_inverted_index.early | 92 |
| abstract_inverted_index.human | 115 |
| abstract_inverted_index.major | 218 |
| abstract_inverted_index.might | 243 |
| abstract_inverted_index.novel | 37 |
| abstract_inverted_index.order | 21 |
| abstract_inverted_index.organ | 219 |
| abstract_inverted_index.ven-R | 123, 130 |
| abstract_inverted_index.which | 161, 197 |
| abstract_inverted_index.while | 91 |
| abstract_inverted_index.BCL-xL | 149, 190 |
| abstract_inverted_index.First, | 152 |
| abstract_inverted_index.NF-κB | 172 |
| abstract_inverted_index.T-ALL. | 47, 62, 234 |
| abstract_inverted_index.cells, | 71, 124 |
| abstract_inverted_index.cells. | 133 |
| abstract_inverted_index.higher | 97 |
| abstract_inverted_index.kinase | 241 |
| abstract_inverted_index.lines, | 160 |
| abstract_inverted_index.looked | 29 |
| abstract_inverted_index.model, | 209 |
| abstract_inverted_index.showed | 118 |
| abstract_inverted_index.single | 41 |
| abstract_inverted_index.(NWP-R) | 70 |
| abstract_inverted_index.(T-ALL) | 5 |
| abstract_inverted_index.(ven-R) | 67 |
| abstract_inverted_index.BCL-xL, | 90 |
| abstract_inverted_index.BCL-xL. | 178 |
| abstract_inverted_index.T-acute | 2 |
| abstract_inverted_index.Typical | 83 |
| abstract_inverted_index.against | 107, 246 |
| abstract_inverted_index.develop | 23 |
| abstract_inverted_index.drivers | 143 |
| abstract_inverted_index.limited | 7 |
| abstract_inverted_index.mimetic | 59, 231 |
| abstract_inverted_index.pathway | 173 |
| abstract_inverted_index.pre-TCR | 137 |
| abstract_inverted_index.synergy | 216 |
| abstract_inverted_index.therapy | 45 |
| abstract_inverted_index.without | 217 |
| abstract_inverted_index.Methods: | 48 |
| abstract_inverted_index.NWP-0476 | 212 |
| abstract_inverted_index.Relapsed | 1 |
| abstract_inverted_index.Results: | 82 |
| abstract_inverted_index.activity | 121, 128 |
| abstract_inverted_index.control, | 65 |
| abstract_inverted_index.critical | 227 |
| abstract_inverted_index.efficacy | 33 |
| abstract_inverted_index.function | 201 |
| abstract_inverted_index.identify | 77 |
| abstract_inverted_index.isogenic | 64 |
| abstract_inverted_index.leukemia | 4 |
| abstract_inverted_index.mimetics | 17, 238 |
| abstract_inverted_index.options. | 9 |
| abstract_inverted_index.pathway, | 196 |
| abstract_inverted_index.pathways | 141, 225 |
| abstract_inverted_index.rational | 24 |
| abstract_inverted_index.relapsed | 247 |
| abstract_inverted_index.response | 60 |
| abstract_inverted_index.resulted | 162, 182 |
| abstract_inverted_index.silenced | 154 |
| abstract_inverted_index.subtypes | 109 |
| abstract_inverted_index.tyrosine | 240 |
| abstract_inverted_index.(ETP)-ALL | 94 |
| abstract_inverted_index.NWP-0476, | 35 |
| abstract_inverted_index.Silencing | 179 |
| abstract_inverted_index.T-lineage | 111 |
| abstract_inverted_index.activated | 171 |
| abstract_inverted_index.dasatinib | 214 |
| abstract_inverted_index.effective | 106, 245 |
| abstract_inverted_index.increased | 87, 119, 126, 164, 184 |
| abstract_inverted_index.inhibited | 198 |
| abstract_inverted_index.pathways. | 81 |
| abstract_inverted_index.performed | 75 |
| abstract_inverted_index.profiling | 52 |
| abstract_inverted_index.regulated | 79 |
| abstract_inverted_index.signaling | 80, 140, 193, 224 |
| abstract_inverted_index.survival. | 101 |
| abstract_inverted_index.toxicity. | 220 |
| abstract_inverted_index.treatment | 8 |
| abstract_inverted_index.xenograft | 208 |
| abstract_inverted_index.71-protein | 114 |
| abstract_inverted_index.dependence | 88, 99 |
| abstract_inverted_index.expression | 176 |
| abstract_inverted_index.inhibitor, | 39 |
| abstract_inverted_index.inhibitors | 104, 242 |
| abstract_inverted_index.mechanisms | 12 |
| abstract_inverted_index.predictive | 55 |
| abstract_inverted_index.regulators | 228 |
| abstract_inverted_index.resistance | 14, 145, 232 |
| abstract_inverted_index.Combination | 235 |
| abstract_inverted_index.T-precursor | 93 |
| abstract_inverted_index.combination | 25, 44, 210 |
| abstract_inverted_index.inhibition, | 150 |
| abstract_inverted_index.inhibition. | 168 |
| abstract_inverted_index.inhibitors. | 191 |
| abstract_inverted_index.preclinical | 32 |
| abstract_inverted_index.sensitivity | 165, 185 |
| abstract_inverted_index.strategies. | 26 |
| abstract_inverted_index.BCL-2/BCL-xL | 38, 102 |
| abstract_inverted_index.demonstrated | 215 |
| abstract_inverted_index.hypothesized | 135 |
| abstract_inverted_index.investigated | 11 |
| abstract_inverted_index.up-regulated | 194 |
| abstract_inverted_index.lymphoblastic | 3 |
| abstract_inverted_index.phosphokinase | 72, 116 |
| abstract_inverted_index.pro-apoptotic | 200 |
| abstract_inverted_index.respectively. | 151 |
| abstract_inverted_index.differentially | 78 |
| abstract_inverted_index.Conclusions:LCK | 221 |
| abstract_inverted_index.patient-derived | 207 |
| abstract_inverted_index.Mechanistically, | 169 |
| abstract_inverted_index.NWP-0476-resistant | 69 |
| abstract_inverted_index.venetoclax-resistant | 66 |
| abstract_inverted_index.T-ALL.</p></div> | 248 |
| abstract_inverted_index.<div>Abstract<p>Purpose: | 0 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 20 |
| citation_normalized_percentile.value | 0.27448317 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |