Dataset supporting the paper "Multilocus Sequence Typing (MLST) of Wolbachia strains infecting different Culex mosquito species in Côte d'Ivoire" Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.17669477
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.17669477
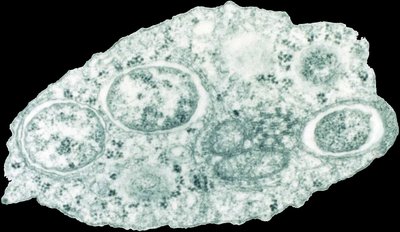
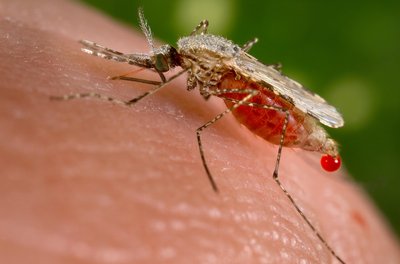
These data accompany the publication “Multilocus Sequence Typing (MLST) of Wolbachia strains infecting different Culex mosquito species in Côte d’Ivoire”They consist of three files, which are as follows: COII gene sequences of Culex mosquito species collected in Côte d'Ivoire. Alignment of coxA gene sequences of Wolbachia infected Culex mosquito species caught in Côte d'Ivoire. Alignment of fbpA gene sequences of Wolbachia infected Culex mosquito species caught in Côte d'Ivoire. Alignment of gatB gene sequences of Wolbachia infected Culex mosquito species caught in Côte d'Ivoire. Alignment of ftsZ gene sequences of Wolbachia infected Culex mosquito species caught in Côte d'Ivoire. Alignnment of MLST concatenated sequences. Alignment of hcpA gene sequences of Wolbachia infected Culex mosquito species caught in Côte d'Ivoire. The database generated in this study contains information on the morphological identification of Culex mosquito species, the mosquito collection sites, and data related to the natural infection of Culex species by Wolbachia. Statistic analysis performed in R cifA gene sequences of Wolbachia strains hosted by Culex mosquitoes collected in Côte d’Ivoire.
Related Topics
- Type
- dataset
- Landing Page
- https://doi.org/10.5281/zenodo.17669477
- OA Status
- green
- OpenAlex ID
- https://openalex.org/W7106336131
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W7106336131Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.5281/zenodo.17669477Digital Object Identifier
- Title
-
Dataset supporting the paper "Multilocus Sequence Typing (MLST) of Wolbachia strains infecting different Culex mosquito species in Côte d'Ivoire"Work title
- Type
-
datasetOpenAlex work type
- Publication year
-
2025Year of publication
- Publication date
-
2025-11-25Full publication date if available
- Authors
-
YAO, Raymond Karlhis, Bilgo Etienne, GOMGNIMBOU, Michel KiréoporiList of authors in order
- Landing page
-
https://doi.org/10.5281/zenodo.17669477Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.5281/zenodo.17669477Direct OA link when available
- Concepts
-
Wolbachia, Biology, Culex, Multilocus sequence typing, Genetics, Culex pipiens, Gene, Virology, Phylogenetics, Zoology, Sequence analysis, Phylogenetic tree, Anopheles, Typing, DNA sequencing, Mitochondrial DNATop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
Full payload
| id | https://openalex.org/W7106336131 |
|---|---|
| doi | https://doi.org/10.5281/zenodo.17669477 |
| ids.doi | https://doi.org/10.5281/zenodo.17669477 |
| ids.openalex | https://openalex.org/W7106336131 |
| fwci | |
| type | dataset |
| title | Dataset supporting the paper "Multilocus Sequence Typing (MLST) of Wolbachia strains infecting different Culex mosquito species in Côte d'Ivoire" |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2779356413 |
| concepts[0].level | 3 |
| concepts[0].score | 0.9571974873542786 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q283526 |
| concepts[0].display_name | Wolbachia |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.8565024733543396 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C2779518617 |
| concepts[2].level | 3 |
| concepts[2].score | 0.8028623461723328 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q276595 |
| concepts[2].display_name | Culex |
| concepts[3].id | https://openalex.org/C77434933 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5170274376869202 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q4043440 |
| concepts[3].display_name | Multilocus sequence typing |
| concepts[4].id | https://openalex.org/C54355233 |
| concepts[4].level | 1 |
| concepts[4].score | 0.4570220112800598 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[4].display_name | Genetics |
| concepts[5].id | https://openalex.org/C2776442642 |
| concepts[5].level | 3 |
| concepts[5].score | 0.42878592014312744 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q27958 |
| concepts[5].display_name | Culex pipiens |
| concepts[6].id | https://openalex.org/C104317684 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4199880063533783 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[6].display_name | Gene |
| concepts[7].id | https://openalex.org/C159047783 |
| concepts[7].level | 1 |
| concepts[7].score | 0.3952977657318115 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7215 |
| concepts[7].display_name | Virology |
| concepts[8].id | https://openalex.org/C90132467 |
| concepts[8].level | 3 |
| concepts[8].score | 0.3541053831577301 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q171184 |
| concepts[8].display_name | Phylogenetics |
| concepts[9].id | https://openalex.org/C90856448 |
| concepts[9].level | 1 |
| concepts[9].score | 0.35023233294487 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q431 |
| concepts[9].display_name | Zoology |
| concepts[10].id | https://openalex.org/C61053724 |
| concepts[10].level | 3 |
| concepts[10].score | 0.3407740890979767 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q1154615 |
| concepts[10].display_name | Sequence analysis |
| concepts[11].id | https://openalex.org/C193252679 |
| concepts[11].level | 3 |
| concepts[11].score | 0.3140285909175873 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q242125 |
| concepts[11].display_name | Phylogenetic tree |
| concepts[12].id | https://openalex.org/C2781375639 |
| concepts[12].level | 3 |
| concepts[12].score | 0.2934817969799042 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q158597 |
| concepts[12].display_name | Anopheles |
| concepts[13].id | https://openalex.org/C2781209916 |
| concepts[13].level | 2 |
| concepts[13].score | 0.2782149314880371 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q3215756 |
| concepts[13].display_name | Typing |
| concepts[14].id | https://openalex.org/C51679486 |
| concepts[14].level | 3 |
| concepts[14].score | 0.2716275155544281 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q380546 |
| concepts[14].display_name | DNA sequencing |
| concepts[15].id | https://openalex.org/C24586158 |
| concepts[15].level | 3 |
| concepts[15].score | 0.25248464941978455 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q27075 |
| concepts[15].display_name | Mitochondrial DNA |
| keywords[0].id | https://openalex.org/keywords/wolbachia |
| keywords[0].score | 0.9571974873542786 |
| keywords[0].display_name | Wolbachia |
| keywords[1].id | https://openalex.org/keywords/culex |
| keywords[1].score | 0.8028623461723328 |
| keywords[1].display_name | Culex |
| keywords[2].id | https://openalex.org/keywords/multilocus-sequence-typing |
| keywords[2].score | 0.5170274376869202 |
| keywords[2].display_name | Multilocus sequence typing |
| keywords[3].id | https://openalex.org/keywords/culex-pipiens |
| keywords[3].score | 0.42878592014312744 |
| keywords[3].display_name | Culex pipiens |
| keywords[4].id | https://openalex.org/keywords/gene |
| keywords[4].score | 0.4199880063533783 |
| keywords[4].display_name | Gene |
| keywords[5].id | https://openalex.org/keywords/phylogenetics |
| keywords[5].score | 0.3541053831577301 |
| keywords[5].display_name | Phylogenetics |
| language | |
| locations[0].id | doi:10.5281/zenodo.17669477 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400562 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| locations[0].source.host_organization | https://openalex.org/I67311998 |
| locations[0].source.host_organization_name | European Organization for Nuclear Research |
| locations[0].source.host_organization_lineage | https://openalex.org/I67311998 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | dataset |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.5281/zenodo.17669477 |
| indexed_in | datacite |
| authorships[0].author.id | |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | YAO, Raymond Karlhis |
| authorships[0].countries | BF |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210156996 |
| authorships[0].affiliations[0].raw_affiliation_string | Equipe Biologie Moléculaire et Biotechnologies, Laboratoire de Recherche, Centre MURAZ, Institut National de Santé Publique, Bobo-Dioulasso, Burkina Faso |
| authorships[0].institutions[0].id | https://openalex.org/I4210156996 |
| authorships[0].institutions[0].ror | https://ror.org/https://ror.org/04nhm0g90 |
| authorships[0].institutions[0].type | other |
| authorships[0].institutions[0].lineage | https://openalex.org/I4210156996 |
| authorships[0].institutions[0].country_code | BF |
| authorships[0].institutions[0].display_name | Centre Muraz |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | YAO, Raymond Karlhis |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Equipe Biologie Moléculaire et Biotechnologies, Laboratoire de Recherche, Centre MURAZ, Institut National de Santé Publique, Bobo-Dioulasso, Burkina Faso |
| authorships[1].author.id | https://openalex.org/A3189310099 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Bilgo Etienne |
| authorships[1].countries | BF |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210163846 |
| authorships[1].affiliations[0].raw_affiliation_string | Institut de Recherche en Sciences de la Santé (IRSS), Direction Régionale de l'Ouest, Bobo-Dioulasso, Burkina Faso |
| authorships[1].institutions[0].id | https://openalex.org/I4210163846 |
| authorships[1].institutions[0].ror | https://ror.org/https://ror.org/05m88q091 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I4210116536, https://openalex.org/I4210163846 |
| authorships[1].institutions[0].country_code | BF |
| authorships[1].institutions[0].display_name | Institut de Recherche en Sciences de la Santé |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | BILGO, Etienne |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Institut de Recherche en Sciences de la Santé (IRSS), Direction Régionale de l'Ouest, Bobo-Dioulasso, Burkina Faso |
| authorships[2].author.id | |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | GOMGNIMBOU, Michel Kiréopori |
| authorships[2].countries | BF |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210156996 |
| authorships[2].affiliations[0].raw_affiliation_string | Equipe Biologie Moléculaire et Biotechnologies, Laboratoire de Recherche, Centre MURAZ, Institut National de Santé Publique, Bobo-Dioulasso, Burkina Faso |
| authorships[2].institutions[0].id | https://openalex.org/I4210156996 |
| authorships[2].institutions[0].ror | https://ror.org/https://ror.org/04nhm0g90 |
| authorships[2].institutions[0].type | other |
| authorships[2].institutions[0].lineage | https://openalex.org/I4210156996 |
| authorships[2].institutions[0].country_code | BF |
| authorships[2].institutions[0].display_name | Centre Muraz |
| authorships[2].author_position | last |
| authorships[2].raw_author_name | GOMGNIMBOU, Michel Kiréopori |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Equipe Biologie Moléculaire et Biotechnologies, Laboratoire de Recherche, Centre MURAZ, Institut National de Santé Publique, Bobo-Dioulasso, Burkina Faso |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.5281/zenodo.17669477 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-11-23T00:00:00 |
| display_name | Dataset supporting the paper "Multilocus Sequence Typing (MLST) of Wolbachia strains infecting different Culex mosquito species in Côte d'Ivoire" |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-27T01:12:40.094763 |
| primary_topic | |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.5281/zenodo.17669477 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400562 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| best_oa_location.source.host_organization | https://openalex.org/I67311998 |
| best_oa_location.source.host_organization_name | European Organization for Nuclear Research |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | dataset |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.5281/zenodo.17669477 |
| primary_location.id | doi:10.5281/zenodo.17669477 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400562 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| primary_location.source.host_organization | https://openalex.org/I67311998 |
| primary_location.source.host_organization_name | European Organization for Nuclear Research |
| primary_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | dataset |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.5281/zenodo.17669477 |
| publication_date | 2025-11-25 |
| publication_year | 2025 |
| referenced_works_count | 0 |
| abstract_inverted_index.R | 155 |
| abstract_inverted_index.as | 26 |
| abstract_inverted_index.by | 149, 163 |
| abstract_inverted_index.in | 17, 36, 51, 66, 81, 96, 116, 122, 154, 167 |
| abstract_inverted_index.of | 9, 21, 31, 40, 44, 55, 59, 70, 74, 85, 89, 100, 105, 109, 131, 146, 159 |
| abstract_inverted_index.on | 127 |
| abstract_inverted_index.to | 142 |
| abstract_inverted_index.The | 119 |
| abstract_inverted_index.and | 139 |
| abstract_inverted_index.are | 25 |
| abstract_inverted_index.the | 3, 128, 135, 143 |
| abstract_inverted_index.COII | 28 |
| abstract_inverted_index.MLST | 101 |
| abstract_inverted_index.cifA | 156 |
| abstract_inverted_index.coxA | 41 |
| abstract_inverted_index.data | 1, 140 |
| abstract_inverted_index.fbpA | 56 |
| abstract_inverted_index.ftsZ | 86 |
| abstract_inverted_index.gatB | 71 |
| abstract_inverted_index.gene | 29, 42, 57, 72, 87, 107, 157 |
| abstract_inverted_index.hcpA | 106 |
| abstract_inverted_index.this | 123 |
| abstract_inverted_index.Culex | 14, 32, 47, 62, 77, 92, 112, 132, 147, 164 |
| abstract_inverted_index.Côte | 18, 37, 52, 67, 82, 97, 117, 168 |
| abstract_inverted_index.These | 0 |
| abstract_inverted_index.study | 124 |
| abstract_inverted_index.three | 22 |
| abstract_inverted_index.which | 24 |
| abstract_inverted_index.(MLST) | 8 |
| abstract_inverted_index.Typing | 7 |
| abstract_inverted_index.caught | 50, 65, 80, 95, 115 |
| abstract_inverted_index.files, | 23 |
| abstract_inverted_index.hosted | 162 |
| abstract_inverted_index.sites, | 138 |
| abstract_inverted_index.consist | 20 |
| abstract_inverted_index.natural | 144 |
| abstract_inverted_index.related | 141 |
| abstract_inverted_index.species | 16, 34, 49, 64, 79, 94, 114, 148 |
| abstract_inverted_index.strains | 11, 161 |
| abstract_inverted_index.Sequence | 6 |
| abstract_inverted_index.analysis | 152 |
| abstract_inverted_index.contains | 125 |
| abstract_inverted_index.database | 120 |
| abstract_inverted_index.follows: | 27 |
| abstract_inverted_index.infected | 46, 61, 76, 91, 111 |
| abstract_inverted_index.mosquito | 15, 33, 48, 63, 78, 93, 113, 133, 136 |
| abstract_inverted_index.species, | 134 |
| abstract_inverted_index.Alignment | 39, 54, 69, 84, 104 |
| abstract_inverted_index.Statistic | 151 |
| abstract_inverted_index.Wolbachia | 10, 45, 60, 75, 90, 110, 160 |
| abstract_inverted_index.accompany | 2 |
| abstract_inverted_index.collected | 35, 166 |
| abstract_inverted_index.d'Ivoire. | 38, 53, 68, 83, 98, 118 |
| abstract_inverted_index.different | 13 |
| abstract_inverted_index.generated | 121 |
| abstract_inverted_index.infecting | 12 |
| abstract_inverted_index.infection | 145 |
| abstract_inverted_index.performed | 153 |
| abstract_inverted_index.sequences | 30, 43, 58, 73, 88, 108, 158 |
| abstract_inverted_index.Alignnment | 99 |
| abstract_inverted_index.Wolbachia. | 150 |
| abstract_inverted_index.collection | 137 |
| abstract_inverted_index.mosquitoes | 165 |
| abstract_inverted_index.sequences. | 103 |
| abstract_inverted_index.d’Ivoire. | 169 |
| abstract_inverted_index.information | 126 |
| abstract_inverted_index.publication | 4 |
| abstract_inverted_index.concatenated | 102 |
| abstract_inverted_index.morphological | 129 |
| abstract_inverted_index.“Multilocus | 5 |
| abstract_inverted_index.identification | 130 |
| abstract_inverted_index.d’Ivoire”They | 19 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 3 |
| citation_normalized_percentile |