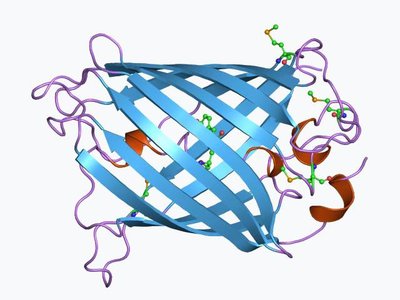
Design And Cloning Of A Pair Of Split-Gfp Constructs To Examine One Of The Split-Sites For Scn1A Article Swipe
Wen Yih Aw
,
David Morris
·
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.1473858
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.1473858
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.1473858
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.1473858
Design and cloning of split-GFP constructs to investigate a potential split-sites for SCN1A fragments complementation.
Related Topics
Concepts
Metadata
- Type
- dataset
- Language
- en
- Landing Page
- https://doi.org/10.5281/zenodo.1473858
- OA Status
- green
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4394036825
All OpenAlex metadata
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4394036825Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.5281/zenodo.1473858Digital Object Identifier
- Title
-
Design And Cloning Of A Pair Of Split-Gfp Constructs To Examine One Of The Split-Sites For Scn1AWork title
- Type
-
datasetOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2018Year of publication
- Publication date
-
2018-10-30Full publication date if available
- Authors
-
Wen Yih Aw, David MorrisList of authors in order
- Landing page
-
https://doi.org/10.5281/zenodo.1473858Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.5281/zenodo.1473858Direct OA link when available
- Concepts
-
Cloning (programming), Computational biology, Computer science, Green fluorescent protein, Biology, Genetics, Theoretical computer science, Programming language, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4394036825 |
|---|---|
| doi | https://doi.org/10.5281/zenodo.1473858 |
| ids.doi | https://doi.org/10.5281/zenodo.1473858 |
| ids.openalex | https://openalex.org/W4394036825 |
| fwci | |
| type | dataset |
| title | Design And Cloning Of A Pair Of Split-Gfp Constructs To Examine One Of The Split-Sites For Scn1A |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11032 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.8352000117301941 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1708 |
| topics[0].subfield.display_name | Hardware and Architecture |
| topics[0].display_name | VLSI and Analog Circuit Testing |
| topics[1].id | https://openalex.org/T14117 |
| topics[1].field.id | https://openalex.org/fields/22 |
| topics[1].field.display_name | Engineering |
| topics[1].score | 0.7439000010490417 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2208 |
| topics[1].subfield.display_name | Electrical and Electronic Engineering |
| topics[1].display_name | Integrated Circuits and Semiconductor Failure Analysis |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C121050878 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7170677781105042 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q5135020 |
| concepts[0].display_name | Cloning (programming) |
| concepts[1].id | https://openalex.org/C70721500 |
| concepts[1].level | 1 |
| concepts[1].score | 0.5619361400604248 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[1].display_name | Computational biology |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.45000413060188293 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C142613039 |
| concepts[3].level | 3 |
| concepts[3].score | 0.4157867431640625 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q272983 |
| concepts[3].display_name | Green fluorescent protein |
| concepts[4].id | https://openalex.org/C86803240 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4095669090747833 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[4].display_name | Biology |
| concepts[5].id | https://openalex.org/C54355233 |
| concepts[5].level | 1 |
| concepts[5].score | 0.36487680673599243 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[5].display_name | Genetics |
| concepts[6].id | https://openalex.org/C80444323 |
| concepts[6].level | 1 |
| concepts[6].score | 0.3410780131816864 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q2878974 |
| concepts[6].display_name | Theoretical computer science |
| concepts[7].id | https://openalex.org/C199360897 |
| concepts[7].level | 1 |
| concepts[7].score | 0.1952640414237976 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[7].display_name | Programming language |
| concepts[8].id | https://openalex.org/C104317684 |
| concepts[8].level | 2 |
| concepts[8].score | 0.094952791929245 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[8].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/cloning |
| keywords[0].score | 0.7170677781105042 |
| keywords[0].display_name | Cloning (programming) |
| keywords[1].id | https://openalex.org/keywords/computational-biology |
| keywords[1].score | 0.5619361400604248 |
| keywords[1].display_name | Computational biology |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.45000413060188293 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/green-fluorescent-protein |
| keywords[3].score | 0.4157867431640625 |
| keywords[3].display_name | Green fluorescent protein |
| keywords[4].id | https://openalex.org/keywords/biology |
| keywords[4].score | 0.4095669090747833 |
| keywords[4].display_name | Biology |
| keywords[5].id | https://openalex.org/keywords/genetics |
| keywords[5].score | 0.36487680673599243 |
| keywords[5].display_name | Genetics |
| keywords[6].id | https://openalex.org/keywords/theoretical-computer-science |
| keywords[6].score | 0.3410780131816864 |
| keywords[6].display_name | Theoretical computer science |
| keywords[7].id | https://openalex.org/keywords/programming-language |
| keywords[7].score | 0.1952640414237976 |
| keywords[7].display_name | Programming language |
| keywords[8].id | https://openalex.org/keywords/gene |
| keywords[8].score | 0.094952791929245 |
| keywords[8].display_name | Gene |
| language | en |
| locations[0].id | doi:10.5281/zenodo.1473858 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400562 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| locations[0].source.host_organization | https://openalex.org/I67311998 |
| locations[0].source.host_organization_name | European Organization for Nuclear Research |
| locations[0].source.host_organization_lineage | https://openalex.org/I67311998 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | dataset |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.5281/zenodo.1473858 |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5083456746 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-1724-412X |
| authorships[0].author.display_name | Wen Yih Aw |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I114027177 |
| authorships[0].affiliations[0].raw_affiliation_string | Catalyst for Rare Diseases, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill. |
| authorships[0].institutions[0].id | https://openalex.org/I114027177 |
| authorships[0].institutions[0].ror | https://ror.org/0130frc33 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I114027177 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | University of North Carolina at Chapel Hill |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Wen Yih Aw |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Catalyst for Rare Diseases, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill. |
| authorships[1].author.id | https://openalex.org/A5018464813 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-5542-428X |
| authorships[1].author.display_name | David Morris |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I114027177 |
| authorships[1].affiliations[0].raw_affiliation_string | Catalyst for Rare Diseases, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill. |
| authorships[1].institutions[0].id | https://openalex.org/I114027177 |
| authorships[1].institutions[0].ror | https://ror.org/0130frc33 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I114027177 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of North Carolina at Chapel Hill |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | David Morris |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Catalyst for Rare Diseases, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.5281/zenodo.1473858 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Design And Cloning Of A Pair Of Split-Gfp Constructs To Examine One Of The Split-Sites For Scn1A |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T11032 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.8352000117301941 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1708 |
| primary_topic.subfield.display_name | Hardware and Architecture |
| primary_topic.display_name | VLSI and Analog Circuit Testing |
| related_works | https://openalex.org/W4391375266, https://openalex.org/W4247151890, https://openalex.org/W2025578726, https://openalex.org/W3039663660, https://openalex.org/W2041840213, https://openalex.org/W2061068107, https://openalex.org/W1908246831, https://openalex.org/W1533688586, https://openalex.org/W1969909463, https://openalex.org/W147536002 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.5281/zenodo.1473858 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400562 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| best_oa_location.source.host_organization | https://openalex.org/I67311998 |
| best_oa_location.source.host_organization_name | European Organization for Nuclear Research |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | dataset |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.5281/zenodo.1473858 |
| primary_location.id | doi:10.5281/zenodo.1473858 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400562 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| primary_location.source.host_organization | https://openalex.org/I67311998 |
| primary_location.source.host_organization_name | European Organization for Nuclear Research |
| primary_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | dataset |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.5281/zenodo.1473858 |
| publication_date | 2018-10-30 |
| publication_year | 2018 |
| referenced_works_count | 0 |
| abstract_inverted_index.a | 8 |
| abstract_inverted_index.of | 3 |
| abstract_inverted_index.to | 6 |
| abstract_inverted_index.and | 1 |
| abstract_inverted_index.for | 11 |
| abstract_inverted_index.SCN1A | 12 |
| abstract_inverted_index.Design | 0 |
| abstract_inverted_index.cloning | 2 |
| abstract_inverted_index.fragments | 13 |
| abstract_inverted_index.potential | 9 |
| abstract_inverted_index.split-GFP | 4 |
| abstract_inverted_index.constructs | 5 |
| abstract_inverted_index.investigate | 7 |
| abstract_inverted_index.split-sites | 10 |
| abstract_inverted_index.complementation. | 14 |
| cited_by_percentile_year | |
| countries_distinct_count | 1 |
| institutions_distinct_count | 2 |
| citation_normalized_percentile |