Design of parallel 𝛽‐sheet nanofibrils using Monte Carlo search, coarse‐grained simulations, and experimental testing Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1002/pro.5102
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1002/pro.5102
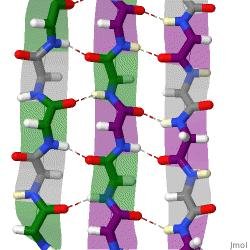
Peptide self‐assembly into amyloid fibrils provides numerous applications in drug delivery and biomedical engineering applications. We augment our previously‐established computational screening technique along with experimental biophysical characterization to discover 7‐mer peptides that self‐assemble into “parallel β ‐sheets”, that is, β ‐sheets with N‐terminus‐to‐C‐terminus 𝛽‐strand vectors oriented in parallel. To accomplish the desired β ‐strand organization, we applied the PepAD amino acid sequence design software to the Class‐1 cross‐ β spine defined by Sawaya et al. This molecular configuration includes two layers of parallel β ‐sheets stacked such that N‐terminus‐to‐C‐terminus vectors are oriented antiparallel for molecules on adjacent β ‐sheets. The first cohort of PepAD identified peptides were examined for their fibrillation behavior in DMD/PRIME20 simulations, and the top performing sequence was selected as a prototype for a subsequent round of sequence refinement. The two rounds of design resulted in a library of eight 7‐mer peptides. In DMD/PRIME20 simulations, five of these peptides spontaneously formed fibril‐like structures with a predominantly parallel 𝛽‐sheet arrangement, two formed fibril‐like structure with <50% in parallel 𝛽‐sheet arrangement and one remained a random coil. Among the eight candidate peptides produced by PepAD and DMD/PRIME20, five were synthesized and purified. All five assembled into amyloid fibrils composed of parallel β ‐sheets based on Fourier transform infrared spectroscopy, circular dichroism, electron microscopy, and thioflavin‐T fluorescence spectroscopy measurements.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1002/pro.5102
- https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102
- OA Status
- hybrid
- Cited By
- 4
- References
- 52
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4400886670
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4400886670Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1002/pro.5102Digital Object Identifier
- Title
-
Design of parallel 𝛽‐sheet nanofibrils using Monte Carlo search, coarse‐grained simulations, and experimental testingWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-07-22Full publication date if available
- Authors
-
Sudeep Sarma, Tarunya Rao Sudarshan, Van Thuan Nguyen, Alicia S. Robang, Xingqing Xiao, Justin V. Le, Michael E. Helmicki, Anant K. Paravastu, Carol K. HallList of authors in order
- Landing page
-
https://doi.org/10.1002/pro.5102Publisher landing page
- PDF URL
-
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102Direct OA link when available
- Concepts
-
Antiparallel (mathematics), Beta sheet, Thioflavin, Random coil, Fibril, Peptide sequence, Peptide, Circular dichroism, Protein secondary structure, Sequence (biology), Chemistry, Crystallography, Materials science, Biophysics, Physics, Biochemistry, Biology, Disease, Quantum mechanics, Alzheimer's disease, Magnetic field, Pathology, Medicine, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
4Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 1Per-year citation counts (last 5 years)
- References (count)
-
52Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4400886670 |
|---|---|
| doi | https://doi.org/10.1002/pro.5102 |
| ids.doi | https://doi.org/10.1002/pro.5102 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39037281 |
| ids.openalex | https://openalex.org/W4400886670 |
| fwci | 1.40401299 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000072757 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Protein Conformation, beta-Strand |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D009010 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Monte Carlo Method |
| mesh[2].qualifier_ui | Q000737 |
| mesh[2].descriptor_ui | D057139 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | chemistry |
| mesh[2].descriptor_name | Nanofibers |
| mesh[3].qualifier_ui | Q000737 |
| mesh[3].descriptor_ui | D010455 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | chemistry |
| mesh[3].descriptor_name | Peptides |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D000595 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Amino Acid Sequence |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D017433 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Protein Structure, Secondary |
| mesh[6].qualifier_ui | Q000737 |
| mesh[6].descriptor_ui | D000682 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | chemistry |
| mesh[6].descriptor_name | Amyloid |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D008958 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Models, Molecular |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D056004 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Molecular Dynamics Simulation |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D000072757 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Protein Conformation, beta-Strand |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D009010 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Monte Carlo Method |
| mesh[11].qualifier_ui | Q000737 |
| mesh[11].descriptor_ui | D057139 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | chemistry |
| mesh[11].descriptor_name | Nanofibers |
| mesh[12].qualifier_ui | Q000737 |
| mesh[12].descriptor_ui | D010455 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | chemistry |
| mesh[12].descriptor_name | Peptides |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D000595 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Amino Acid Sequence |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D017433 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Protein Structure, Secondary |
| mesh[15].qualifier_ui | Q000737 |
| mesh[15].descriptor_ui | D000682 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | chemistry |
| mesh[15].descriptor_name | Amyloid |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D008958 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Models, Molecular |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D056004 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Molecular Dynamics Simulation |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D000072757 |
| mesh[18].is_major_topic | True |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Protein Conformation, beta-Strand |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D009010 |
| mesh[19].is_major_topic | True |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Monte Carlo Method |
| mesh[20].qualifier_ui | Q000737 |
| mesh[20].descriptor_ui | D057139 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | chemistry |
| mesh[20].descriptor_name | Nanofibers |
| mesh[21].qualifier_ui | Q000737 |
| mesh[21].descriptor_ui | D010455 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | chemistry |
| mesh[21].descriptor_name | Peptides |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D000595 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Amino Acid Sequence |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D017433 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Protein Structure, Secondary |
| mesh[24].qualifier_ui | Q000737 |
| mesh[24].descriptor_ui | D000682 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | chemistry |
| mesh[24].descriptor_name | Amyloid |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D008958 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Models, Molecular |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D056004 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | Molecular Dynamics Simulation |
| type | article |
| title | Design of parallel 𝛽‐sheet nanofibrils using Monte Carlo search, coarse‐grained simulations, and experimental testing |
| biblio.issue | 8 |
| biblio.volume | 33 |
| biblio.last_page | e5102 |
| biblio.first_page | e5102 |
| topics[0].id | https://openalex.org/T11419 |
| topics[0].field.id | https://openalex.org/fields/25 |
| topics[0].field.display_name | Materials Science |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2502 |
| topics[0].subfield.display_name | Biomaterials |
| topics[0].display_name | Supramolecular Self-Assembly in Materials |
| topics[1].id | https://openalex.org/T14318 |
| topics[1].field.id | https://openalex.org/fields/16 |
| topics[1].field.display_name | Chemistry |
| topics[1].score | 0.9973000288009644 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1605 |
| topics[1].subfield.display_name | Organic Chemistry |
| topics[1].display_name | Polydiacetylene-based materials and applications |
| topics[2].id | https://openalex.org/T10407 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9861000180244446 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Lipid Membrane Structure and Behavior |
| is_xpac | False |
| apc_list.value | 4070 |
| apc_list.currency | USD |
| apc_list.value_usd | 4070 |
| apc_paid.value | 4070 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 4070 |
| concepts[0].id | https://openalex.org/C142089489 |
| concepts[0].level | 3 |
| concepts[0].score | 0.828825056552887 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1053976 |
| concepts[0].display_name | Antiparallel (mathematics) |
| concepts[1].id | https://openalex.org/C159547669 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6400927305221558 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q283449 |
| concepts[1].display_name | Beta sheet |
| concepts[2].id | https://openalex.org/C2778896982 |
| concepts[2].level | 4 |
| concepts[2].score | 0.6122456192970276 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2033625 |
| concepts[2].display_name | Thioflavin |
| concepts[3].id | https://openalex.org/C34349673 |
| concepts[3].level | 3 |
| concepts[3].score | 0.6101665496826172 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q899124 |
| concepts[3].display_name | Random coil |
| concepts[4].id | https://openalex.org/C27523624 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5745497941970825 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q14860849 |
| concepts[4].display_name | Fibril |
| concepts[5].id | https://openalex.org/C167625842 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5019748210906982 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q899763 |
| concepts[5].display_name | Peptide sequence |
| concepts[6].id | https://openalex.org/C2779281246 |
| concepts[6].level | 2 |
| concepts[6].score | 0.48632723093032837 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q172847 |
| concepts[6].display_name | Peptide |
| concepts[7].id | https://openalex.org/C133571119 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4739556312561035 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q899102 |
| concepts[7].display_name | Circular dichroism |
| concepts[8].id | https://openalex.org/C62614982 |
| concepts[8].level | 2 |
| concepts[8].score | 0.47048595547676086 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q904984 |
| concepts[8].display_name | Protein secondary structure |
| concepts[9].id | https://openalex.org/C2778112365 |
| concepts[9].level | 2 |
| concepts[9].score | 0.46863460540771484 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q3511065 |
| concepts[9].display_name | Sequence (biology) |
| concepts[10].id | https://openalex.org/C185592680 |
| concepts[10].level | 0 |
| concepts[10].score | 0.44507041573524475 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[10].display_name | Chemistry |
| concepts[11].id | https://openalex.org/C8010536 |
| concepts[11].level | 1 |
| concepts[11].score | 0.42719462513923645 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q160398 |
| concepts[11].display_name | Crystallography |
| concepts[12].id | https://openalex.org/C192562407 |
| concepts[12].level | 0 |
| concepts[12].score | 0.4161979854106903 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q228736 |
| concepts[12].display_name | Materials science |
| concepts[13].id | https://openalex.org/C12554922 |
| concepts[13].level | 1 |
| concepts[13].score | 0.39478686451911926 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7100 |
| concepts[13].display_name | Biophysics |
| concepts[14].id | https://openalex.org/C121332964 |
| concepts[14].level | 0 |
| concepts[14].score | 0.1657552421092987 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[14].display_name | Physics |
| concepts[15].id | https://openalex.org/C55493867 |
| concepts[15].level | 1 |
| concepts[15].score | 0.14879244565963745 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[15].display_name | Biochemistry |
| concepts[16].id | https://openalex.org/C86803240 |
| concepts[16].level | 0 |
| concepts[16].score | 0.12263205647468567 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[16].display_name | Biology |
| concepts[17].id | https://openalex.org/C2779134260 |
| concepts[17].level | 2 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q12136 |
| concepts[17].display_name | Disease |
| concepts[18].id | https://openalex.org/C62520636 |
| concepts[18].level | 1 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q944 |
| concepts[18].display_name | Quantum mechanics |
| concepts[19].id | https://openalex.org/C502032728 |
| concepts[19].level | 3 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q11081 |
| concepts[19].display_name | Alzheimer's disease |
| concepts[20].id | https://openalex.org/C115260700 |
| concepts[20].level | 2 |
| concepts[20].score | 0.0 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q11408 |
| concepts[20].display_name | Magnetic field |
| concepts[21].id | https://openalex.org/C142724271 |
| concepts[21].level | 1 |
| concepts[21].score | 0.0 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[21].display_name | Pathology |
| concepts[22].id | https://openalex.org/C71924100 |
| concepts[22].level | 0 |
| concepts[22].score | 0.0 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[22].display_name | Medicine |
| concepts[23].id | https://openalex.org/C104317684 |
| concepts[23].level | 2 |
| concepts[23].score | 0.0 |
| concepts[23].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[23].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/antiparallel |
| keywords[0].score | 0.828825056552887 |
| keywords[0].display_name | Antiparallel (mathematics) |
| keywords[1].id | https://openalex.org/keywords/beta-sheet |
| keywords[1].score | 0.6400927305221558 |
| keywords[1].display_name | Beta sheet |
| keywords[2].id | https://openalex.org/keywords/thioflavin |
| keywords[2].score | 0.6122456192970276 |
| keywords[2].display_name | Thioflavin |
| keywords[3].id | https://openalex.org/keywords/random-coil |
| keywords[3].score | 0.6101665496826172 |
| keywords[3].display_name | Random coil |
| keywords[4].id | https://openalex.org/keywords/fibril |
| keywords[4].score | 0.5745497941970825 |
| keywords[4].display_name | Fibril |
| keywords[5].id | https://openalex.org/keywords/peptide-sequence |
| keywords[5].score | 0.5019748210906982 |
| keywords[5].display_name | Peptide sequence |
| keywords[6].id | https://openalex.org/keywords/peptide |
| keywords[6].score | 0.48632723093032837 |
| keywords[6].display_name | Peptide |
| keywords[7].id | https://openalex.org/keywords/circular-dichroism |
| keywords[7].score | 0.4739556312561035 |
| keywords[7].display_name | Circular dichroism |
| keywords[8].id | https://openalex.org/keywords/protein-secondary-structure |
| keywords[8].score | 0.47048595547676086 |
| keywords[8].display_name | Protein secondary structure |
| keywords[9].id | https://openalex.org/keywords/sequence |
| keywords[9].score | 0.46863460540771484 |
| keywords[9].display_name | Sequence (biology) |
| keywords[10].id | https://openalex.org/keywords/chemistry |
| keywords[10].score | 0.44507041573524475 |
| keywords[10].display_name | Chemistry |
| keywords[11].id | https://openalex.org/keywords/crystallography |
| keywords[11].score | 0.42719462513923645 |
| keywords[11].display_name | Crystallography |
| keywords[12].id | https://openalex.org/keywords/materials-science |
| keywords[12].score | 0.4161979854106903 |
| keywords[12].display_name | Materials science |
| keywords[13].id | https://openalex.org/keywords/biophysics |
| keywords[13].score | 0.39478686451911926 |
| keywords[13].display_name | Biophysics |
| keywords[14].id | https://openalex.org/keywords/physics |
| keywords[14].score | 0.1657552421092987 |
| keywords[14].display_name | Physics |
| keywords[15].id | https://openalex.org/keywords/biochemistry |
| keywords[15].score | 0.14879244565963745 |
| keywords[15].display_name | Biochemistry |
| keywords[16].id | https://openalex.org/keywords/biology |
| keywords[16].score | 0.12263205647468567 |
| keywords[16].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1002/pro.5102 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S156919612 |
| locations[0].source.issn | 0961-8368, 1469-896X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0961-8368 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Protein Science |
| locations[0].source.host_organization | https://openalex.org/P4310320595 |
| locations[0].source.host_organization_name | Wiley |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320595 |
| locations[0].source.host_organization_lineage_names | Wiley |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Protein Science |
| locations[0].landing_page_url | https://doi.org/10.1002/pro.5102 |
| locations[1].id | pmid:39037281 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Protein science : a publication of the Protein Society |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39037281 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:11261811 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Protein Sci |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11261811 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5055187342 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-4642-7689 |
| authorships[0].author.display_name | Sudeep Sarma |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I137902535 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[0].institutions[0].id | https://openalex.org/I137902535 |
| authorships[0].institutions[0].ror | https://ror.org/04tj63d06 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I137902535 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | North Carolina State University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Sudeep Sarma |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[1].author.id | https://openalex.org/A5113051161 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Tarunya Rao Sudarshan |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I130701444 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[1].institutions[0].id | https://openalex.org/I130701444 |
| authorships[1].institutions[0].ror | https://ror.org/01zkghx44 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I130701444 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Georgia Institute of Technology |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Tarunya Rao Sudarshan |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[2].author.id | https://openalex.org/A5108761417 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Van Thuan Nguyen |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I137902535 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[2].institutions[0].id | https://openalex.org/I137902535 |
| authorships[2].institutions[0].ror | https://ror.org/04tj63d06 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I137902535 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | North Carolina State University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Van Nguyen |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[3].author.id | https://openalex.org/A5005831557 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-3857-9559 |
| authorships[3].author.display_name | Alicia S. Robang |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I130701444 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[3].institutions[0].id | https://openalex.org/I130701444 |
| authorships[3].institutions[0].ror | https://ror.org/01zkghx44 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I130701444 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | Georgia Institute of Technology |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Alicia S. Robang |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[4].author.id | https://openalex.org/A5087997669 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-3462-3049 |
| authorships[4].author.display_name | Xingqing Xiao |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I137902535 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[4].institutions[0].id | https://openalex.org/I137902535 |
| authorships[4].institutions[0].ror | https://ror.org/04tj63d06 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I137902535 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | North Carolina State University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Xingqing Xiao |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[5].author.id | https://openalex.org/A5061952681 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Justin V. Le |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I130701444 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[5].institutions[0].id | https://openalex.org/I130701444 |
| authorships[5].institutions[0].ror | https://ror.org/01zkghx44 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I130701444 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | Georgia Institute of Technology |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Justin V. Le |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[6].author.id | https://openalex.org/A5060344128 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Michael E. Helmicki |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I130701444 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[6].institutions[0].id | https://openalex.org/I130701444 |
| authorships[6].institutions[0].ror | https://ror.org/01zkghx44 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I130701444 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | Georgia Institute of Technology |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Michael E. Helmicki |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[7].author.id | https://openalex.org/A5062697029 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-7183-1942 |
| authorships[7].author.display_name | Anant K. Paravastu |
| authorships[7].countries | US |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I130701444 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[7].institutions[0].id | https://openalex.org/I130701444 |
| authorships[7].institutions[0].ror | https://ror.org/01zkghx44 |
| authorships[7].institutions[0].type | education |
| authorships[7].institutions[0].lineage | https://openalex.org/I130701444 |
| authorships[7].institutions[0].country_code | US |
| authorships[7].institutions[0].display_name | Georgia Institute of Technology |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Anant K. Paravastu |
| authorships[7].is_corresponding | True |
| authorships[7].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, USA |
| authorships[8].author.id | https://openalex.org/A5057051008 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-7425-587X |
| authorships[8].author.display_name | Carol K. Hall |
| authorships[8].countries | US |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I137902535 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| authorships[8].institutions[0].id | https://openalex.org/I137902535 |
| authorships[8].institutions[0].ror | https://ror.org/04tj63d06 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I137902535 |
| authorships[8].institutions[0].country_code | US |
| authorships[8].institutions[0].display_name | North Carolina State University |
| authorships[8].author_position | last |
| authorships[8].raw_author_name | Carol K. Hall |
| authorships[8].is_corresponding | True |
| authorships[8].raw_affiliation_strings | Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, North Carolina, USA |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102 |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Design of parallel 𝛽‐sheet nanofibrils using Monte Carlo search, coarse‐grained simulations, and experimental testing |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T11419 |
| primary_topic.field.id | https://openalex.org/fields/25 |
| primary_topic.field.display_name | Materials Science |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2502 |
| primary_topic.subfield.display_name | Biomaterials |
| primary_topic.display_name | Supramolecular Self-Assembly in Materials |
| related_works | https://openalex.org/W2011433843, https://openalex.org/W1970329518, https://openalex.org/W1930638136, https://openalex.org/W2068263379, https://openalex.org/W2029863936, https://openalex.org/W4320892511, https://openalex.org/W1973043035, https://openalex.org/W2032227844, https://openalex.org/W1977762397, https://openalex.org/W2407384706 |
| cited_by_count | 4 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 1 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1002/pro.5102 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S156919612 |
| best_oa_location.source.issn | 0961-8368, 1469-896X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0961-8368 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Protein Science |
| best_oa_location.source.host_organization | https://openalex.org/P4310320595 |
| best_oa_location.source.host_organization_name | Wiley |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320595 |
| best_oa_location.source.host_organization_lineage_names | Wiley |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Protein Science |
| best_oa_location.landing_page_url | https://doi.org/10.1002/pro.5102 |
| primary_location.id | doi:10.1002/pro.5102 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S156919612 |
| primary_location.source.issn | 0961-8368, 1469-896X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0961-8368 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Protein Science |
| primary_location.source.host_organization | https://openalex.org/P4310320595 |
| primary_location.source.host_organization_name | Wiley |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320595 |
| primary_location.source.host_organization_lineage_names | Wiley |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/pro.5102 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Protein Science |
| primary_location.landing_page_url | https://doi.org/10.1002/pro.5102 |
| publication_date | 2024-07-22 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2023954079, https://openalex.org/W2082433496, https://openalex.org/W1971121022, https://openalex.org/W3093609761, https://openalex.org/W2101995697, https://openalex.org/W2001148193, https://openalex.org/W3046739948, https://openalex.org/W2141248208, https://openalex.org/W2166344601, https://openalex.org/W2161417492, https://openalex.org/W2151784315, https://openalex.org/W2039030534, https://openalex.org/W2553977609, https://openalex.org/W2023015350, https://openalex.org/W2029863936, https://openalex.org/W1767826187, https://openalex.org/W1991918935, https://openalex.org/W2718362490, https://openalex.org/W2960255956, https://openalex.org/W2104571730, https://openalex.org/W2082501707, https://openalex.org/W2032190222, https://openalex.org/W2121753059, https://openalex.org/W1980482609, https://openalex.org/W2289302043, https://openalex.org/W4239858085, https://openalex.org/W1526832479, https://openalex.org/W1995247791, https://openalex.org/W2074255526, https://openalex.org/W4386191915, https://openalex.org/W4304196012, https://openalex.org/W2160829101, https://openalex.org/W2117537971, https://openalex.org/W3209576331, https://openalex.org/W2000736585, https://openalex.org/W2626325070, https://openalex.org/W4212883768, https://openalex.org/W2051818264, https://openalex.org/W1996779121, https://openalex.org/W2912172518, https://openalex.org/W2802329786, https://openalex.org/W2763359991, https://openalex.org/W2509964843, https://openalex.org/W2895345572, https://openalex.org/W35241519, https://openalex.org/W2155170227, https://openalex.org/W4220893392, https://openalex.org/W4310052035, https://openalex.org/W4200516109, https://openalex.org/W2171271556, https://openalex.org/W2006225923, https://openalex.org/W2045056402 |
| referenced_works_count | 52 |
| abstract_inverted_index.a | 124, 127, 140, 158, 176 |
| abstract_inverted_index.In | 146 |
| abstract_inverted_index.To | 49 |
| abstract_inverted_index.We | 16 |
| abstract_inverted_index.as | 123 |
| abstract_inverted_index.by | 72, 185 |
| abstract_inverted_index.et | 74 |
| abstract_inverted_index.in | 9, 47, 113, 139, 169 |
| abstract_inverted_index.of | 82, 103, 130, 136, 142, 150, 201 |
| abstract_inverted_index.on | 96, 206 |
| abstract_inverted_index.to | 28, 65 |
| abstract_inverted_index.we | 56 |
| abstract_inverted_index.β | 36, 40, 53, 69, 84, 98, 203 |
| abstract_inverted_index.All | 194 |
| abstract_inverted_index.The | 100, 133 |
| abstract_inverted_index.al. | 75 |
| abstract_inverted_index.and | 12, 116, 173, 187, 192, 215 |
| abstract_inverted_index.are | 91 |
| abstract_inverted_index.for | 94, 109, 126 |
| abstract_inverted_index.is, | 39 |
| abstract_inverted_index.one | 174 |
| abstract_inverted_index.our | 18 |
| abstract_inverted_index.the | 51, 58, 66, 117, 180 |
| abstract_inverted_index.top | 118 |
| abstract_inverted_index.two | 80, 134, 163 |
| abstract_inverted_index.was | 121 |
| abstract_inverted_index.This | 76 |
| abstract_inverted_index.acid | 61 |
| abstract_inverted_index.drug | 10 |
| abstract_inverted_index.five | 149, 189, 195 |
| abstract_inverted_index.into | 3, 34, 197 |
| abstract_inverted_index.such | 87 |
| abstract_inverted_index.that | 32, 38, 88 |
| abstract_inverted_index.were | 107, 190 |
| abstract_inverted_index.with | 24, 42, 157, 167 |
| abstract_inverted_index.Among | 179 |
| abstract_inverted_index.PepAD | 59, 104, 186 |
| abstract_inverted_index.along | 23 |
| abstract_inverted_index.amino | 60 |
| abstract_inverted_index.based | 205 |
| abstract_inverted_index.coil. | 178 |
| abstract_inverted_index.eight | 143, 181 |
| abstract_inverted_index.first | 101 |
| abstract_inverted_index.round | 129 |
| abstract_inverted_index.spine | 70 |
| abstract_inverted_index.their | 110 |
| abstract_inverted_index.these | 151 |
| abstract_inverted_index.Sawaya | 73 |
| abstract_inverted_index.cohort | 102 |
| abstract_inverted_index.design | 63, 137 |
| abstract_inverted_index.formed | 154, 164 |
| abstract_inverted_index.layers | 81 |
| abstract_inverted_index.random | 177 |
| abstract_inverted_index.rounds | 135 |
| abstract_inverted_index.<50% | 168 |
| abstract_inverted_index.7‐mer | 30, 144 |
| abstract_inverted_index.Fourier | 207 |
| abstract_inverted_index.Peptide | 1 |
| abstract_inverted_index.amyloid | 4, 198 |
| abstract_inverted_index.applied | 57 |
| abstract_inverted_index.augment | 17 |
| abstract_inverted_index.defined | 71 |
| abstract_inverted_index.desired | 52 |
| abstract_inverted_index.fibrils | 5, 199 |
| abstract_inverted_index.library | 141 |
| abstract_inverted_index.stacked | 86 |
| abstract_inverted_index.vectors | 45, 90 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.adjacent | 97 |
| abstract_inverted_index.behavior | 112 |
| abstract_inverted_index.circular | 211 |
| abstract_inverted_index.composed | 200 |
| abstract_inverted_index.cross‐ | 68 |
| abstract_inverted_index.delivery | 11 |
| abstract_inverted_index.discover | 29 |
| abstract_inverted_index.electron | 213 |
| abstract_inverted_index.examined | 108 |
| abstract_inverted_index.includes | 79 |
| abstract_inverted_index.infrared | 209 |
| abstract_inverted_index.numerous | 7 |
| abstract_inverted_index.oriented | 46, 92 |
| abstract_inverted_index.parallel | 83, 160, 170, 202 |
| abstract_inverted_index.peptides | 31, 106, 152, 183 |
| abstract_inverted_index.produced | 184 |
| abstract_inverted_index.provides | 6 |
| abstract_inverted_index.remained | 175 |
| abstract_inverted_index.resulted | 138 |
| abstract_inverted_index.selected | 122 |
| abstract_inverted_index.sequence | 62, 120, 131 |
| abstract_inverted_index.software | 64 |
| abstract_inverted_index.Class‐1 | 67 |
| abstract_inverted_index.assembled | 196 |
| abstract_inverted_index.candidate | 182 |
| abstract_inverted_index.molecular | 77 |
| abstract_inverted_index.molecules | 95 |
| abstract_inverted_index.parallel. | 48 |
| abstract_inverted_index.peptides. | 145 |
| abstract_inverted_index.prototype | 125 |
| abstract_inverted_index.purified. | 193 |
| abstract_inverted_index.screening | 21 |
| abstract_inverted_index.structure | 166 |
| abstract_inverted_index.technique | 22 |
| abstract_inverted_index.transform | 208 |
| abstract_inverted_index.‐sheets | 41, 85, 204 |
| abstract_inverted_index.‐strand | 54 |
| abstract_inverted_index.accomplish | 50 |
| abstract_inverted_index.biomedical | 13 |
| abstract_inverted_index.dichroism, | 212 |
| abstract_inverted_index.identified | 105 |
| abstract_inverted_index.performing | 119 |
| abstract_inverted_index.structures | 156 |
| abstract_inverted_index.subsequent | 128 |
| abstract_inverted_index.‐sheets. | 99 |
| abstract_inverted_index.DMD/PRIME20 | 114, 147 |
| abstract_inverted_index.arrangement | 172 |
| abstract_inverted_index.biophysical | 26 |
| abstract_inverted_index.engineering | 14 |
| abstract_inverted_index.microscopy, | 214 |
| abstract_inverted_index.refinement. | 132 |
| abstract_inverted_index.synthesized | 191 |
| abstract_inverted_index.“parallel | 35 |
| abstract_inverted_index.DMD/PRIME20, | 188 |
| abstract_inverted_index.antiparallel | 93 |
| abstract_inverted_index.applications | 8 |
| abstract_inverted_index.arrangement, | 162 |
| abstract_inverted_index.experimental | 25 |
| abstract_inverted_index.fibrillation | 111 |
| abstract_inverted_index.fluorescence | 217 |
| abstract_inverted_index.simulations, | 115, 148 |
| abstract_inverted_index.spectroscopy | 218 |
| abstract_inverted_index.𝛽‐sheet | 161, 171 |
| abstract_inverted_index.applications. | 15 |
| abstract_inverted_index.computational | 20 |
| abstract_inverted_index.configuration | 78 |
| abstract_inverted_index.fibril‐like | 155, 165 |
| abstract_inverted_index.measurements. | 219 |
| abstract_inverted_index.organization, | 55 |
| abstract_inverted_index.predominantly | 159 |
| abstract_inverted_index.spectroscopy, | 210 |
| abstract_inverted_index.spontaneously | 153 |
| abstract_inverted_index.‐sheets”, | 37 |
| abstract_inverted_index.𝛽‐strand | 44 |
| abstract_inverted_index.thioflavin‐T | 216 |
| abstract_inverted_index.self‐assemble | 33 |
| abstract_inverted_index.self‐assembly | 2 |
| abstract_inverted_index.characterization | 27 |
| abstract_inverted_index.previously‐established | 19 |
| abstract_inverted_index.N‐terminus‐to‐C‐terminus | 43, 89 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 90 |
| corresponding_author_ids | https://openalex.org/A5062697029, https://openalex.org/A5057051008 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 9 |
| corresponding_institution_ids | https://openalex.org/I130701444, https://openalex.org/I137902535 |
| citation_normalized_percentile.value | 0.72139688 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |