Designing combination therapies with modeling chaperoned machine learning Article Swipe
 YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pcbi.1007158
YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1371/journal.pcbi.1007158
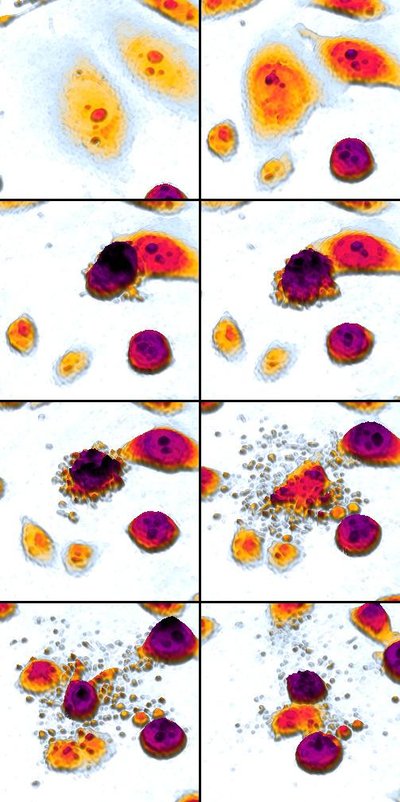
Chemotherapy resistance is a major challenge to the effective treatment of cancer. Thus, a systematic pipeline for the efficient identification of effective combination treatments could bring huge biomedical benefit. In order to facilitate rational design of combination therapies, we developed a comprehensive computational model that incorporates the available biological knowledge and relevant experimental data on the life-and-death response of individual cancer cells to cisplatin or cisplatin combined with the TNF-related apoptosis-inducing ligand (TRAIL). The model's predictions, that a combination treatment of cisplatin and TRAIL would enhance cancer cell death and exhibit a "two-wave killing" temporal pattern, was validated by measuring the dynamics of p53 accumulation, cell fate, and cell death in single cells. The validated model was then subjected to a systematic analysis with an ensemble of diverse machine learning methods. Though each method is characterized by a different algorithm, they collectively identified several molecular players that can sensitize tumor cells to cisplatin-induced apoptosis (sensitizers). The identified sensitizers are consistent with previous experimental observations. Overall, we have illustrated that machine learning analysis of an experimentally validated mechanistic model can convert our available knowledge into the identity of biologically meaningful sensitizers. This knowledge can then be leveraged to design treatment strategies that could improve the efficacy of chemotherapy.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1371/journal.pcbi.1007158
- https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printable
- OA Status
- gold
- Cited By
- 15
- References
- 47
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2972843305
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2972843305Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1371/journal.pcbi.1007158Digital Object Identifier
- Title
-
Designing combination therapies with modeling chaperoned machine learningWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2019Year of publication
- Publication date
-
2019-09-09Full publication date if available
- Authors
-
Yin Zhang, Julie M. Huynh, Guan‐Sheng Liu, Richard Ballweg, Kayenat S. Aryeh, Andrew L. Paek, Tongli ZhangList of authors in order
- Landing page
-
https://doi.org/10.1371/journal.pcbi.1007158Publisher landing page
- PDF URL
-
https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printableDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printableDirect OA link when available
- Concepts
-
Cisplatin, Machine learning, Programmed cell death, Computer science, Pipeline (software), Apoptosis, Rational design, Cancer, Cancer cell, Artificial intelligence, Computational biology, Bioinformatics, Chemotherapy, Biology, Medicine, Nanotechnology, Internal medicine, Materials science, Biochemistry, Programming languageTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
15Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 2, 2023: 1, 2022: 2, 2021: 4Per-year citation counts (last 5 years)
- References (count)
-
47Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2972843305 |
|---|---|
| doi | https://doi.org/10.1371/journal.pcbi.1007158 |
| ids.doi | https://doi.org/10.1371/journal.pcbi.1007158 |
| ids.mag | 2972843305 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/31498788 |
| ids.openalex | https://openalex.org/W2972843305 |
| fwci | 1.92775065 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000465 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Algorithms |
| mesh[1].qualifier_ui | Q000494 |
| mesh[1].descriptor_ui | D000970 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | pharmacology |
| mesh[1].descriptor_name | Antineoplastic Agents |
| mesh[2].qualifier_ui | Q000627 |
| mesh[2].descriptor_ui | D000970 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | therapeutic use |
| mesh[2].descriptor_name | Antineoplastic Agents |
| mesh[3].qualifier_ui | Q000494 |
| mesh[3].descriptor_ui | D002945 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | pharmacology |
| mesh[3].descriptor_name | Cisplatin |
| mesh[4].qualifier_ui | Q000627 |
| mesh[4].descriptor_ui | D002945 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | therapeutic use |
| mesh[4].descriptor_name | Cisplatin |
| mesh[5].qualifier_ui | Q000379 |
| mesh[5].descriptor_ui | D019295 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | methods |
| mesh[5].descriptor_name | Computational Biology |
| mesh[6].qualifier_ui | Q000379 |
| mesh[6].descriptor_ui | D004359 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | methods |
| mesh[6].descriptor_name | Drug Therapy, Combination |
| mesh[7].qualifier_ui | Q000379 |
| mesh[7].descriptor_ui | D004360 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | methods |
| mesh[7].descriptor_name | Drug Therapy, Computer-Assisted |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D006801 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Humans |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D000069550 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Machine Learning |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D008954 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Models, Biological |
| mesh[11].qualifier_ui | Q000188 |
| mesh[11].descriptor_ui | D009369 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | drug therapy |
| mesh[11].descriptor_name | Neoplasms |
| mesh[12].qualifier_ui | Q000187 |
| mesh[12].descriptor_ui | D015398 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | drug effects |
| mesh[12].descriptor_name | Signal Transduction |
| mesh[13].qualifier_ui | Q000494 |
| mesh[13].descriptor_ui | D053221 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | pharmacology |
| mesh[13].descriptor_name | TNF-Related Apoptosis-Inducing Ligand |
| mesh[14].qualifier_ui | Q000627 |
| mesh[14].descriptor_ui | D053221 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | therapeutic use |
| mesh[14].descriptor_name | TNF-Related Apoptosis-Inducing Ligand |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D000465 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Algorithms |
| mesh[16].qualifier_ui | Q000494 |
| mesh[16].descriptor_ui | D000970 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | pharmacology |
| mesh[16].descriptor_name | Antineoplastic Agents |
| mesh[17].qualifier_ui | Q000627 |
| mesh[17].descriptor_ui | D000970 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | therapeutic use |
| mesh[17].descriptor_name | Antineoplastic Agents |
| mesh[18].qualifier_ui | Q000494 |
| mesh[18].descriptor_ui | D002945 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | pharmacology |
| mesh[18].descriptor_name | Cisplatin |
| mesh[19].qualifier_ui | Q000627 |
| mesh[19].descriptor_ui | D002945 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | therapeutic use |
| mesh[19].descriptor_name | Cisplatin |
| mesh[20].qualifier_ui | Q000379 |
| mesh[20].descriptor_ui | D019295 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | methods |
| mesh[20].descriptor_name | Computational Biology |
| mesh[21].qualifier_ui | Q000379 |
| mesh[21].descriptor_ui | D004359 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | methods |
| mesh[21].descriptor_name | Drug Therapy, Combination |
| mesh[22].qualifier_ui | Q000379 |
| mesh[22].descriptor_ui | D004360 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | methods |
| mesh[22].descriptor_name | Drug Therapy, Computer-Assisted |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D006801 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Humans |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D000069550 |
| mesh[24].is_major_topic | True |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Machine Learning |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D008954 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Models, Biological |
| mesh[26].qualifier_ui | Q000188 |
| mesh[26].descriptor_ui | D009369 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | drug therapy |
| mesh[26].descriptor_name | Neoplasms |
| mesh[27].qualifier_ui | Q000187 |
| mesh[27].descriptor_ui | D015398 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | drug effects |
| mesh[27].descriptor_name | Signal Transduction |
| mesh[28].qualifier_ui | Q000494 |
| mesh[28].descriptor_ui | D053221 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | pharmacology |
| mesh[28].descriptor_name | TNF-Related Apoptosis-Inducing Ligand |
| mesh[29].qualifier_ui | Q000627 |
| mesh[29].descriptor_ui | D053221 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | therapeutic use |
| mesh[29].descriptor_name | TNF-Related Apoptosis-Inducing Ligand |
| type | article |
| title | Designing combination therapies with modeling chaperoned machine learning |
| biblio.issue | 9 |
| biblio.volume | 15 |
| biblio.last_page | e1007158 |
| biblio.first_page | e1007158 |
| topics[0].id | https://openalex.org/T10211 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9994999766349792 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1703 |
| topics[0].subfield.display_name | Computational Theory and Mathematics |
| topics[0].display_name | Computational Drug Discovery Methods |
| topics[1].id | https://openalex.org/T10044 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9915000200271606 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Protein Structure and Dynamics |
| topics[2].id | https://openalex.org/T12859 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9855999946594238 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1304 |
| topics[2].subfield.display_name | Biophysics |
| topics[2].display_name | Cell Image Analysis Techniques |
| is_xpac | False |
| apc_list.value | 2655 |
| apc_list.currency | USD |
| apc_list.value_usd | 2655 |
| apc_paid.value | 2655 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2655 |
| concepts[0].id | https://openalex.org/C2778239845 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7992560267448425 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q412415 |
| concepts[0].display_name | Cisplatin |
| concepts[1].id | https://openalex.org/C119857082 |
| concepts[1].level | 1 |
| concepts[1].score | 0.5869607925415039 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[1].display_name | Machine learning |
| concepts[2].id | https://openalex.org/C31573885 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5525416135787964 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q304484 |
| concepts[2].display_name | Programmed cell death |
| concepts[3].id | https://openalex.org/C41008148 |
| concepts[3].level | 0 |
| concepts[3].score | 0.5395189523696899 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[3].display_name | Computer science |
| concepts[4].id | https://openalex.org/C43521106 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5271360874176025 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q2165493 |
| concepts[4].display_name | Pipeline (software) |
| concepts[5].id | https://openalex.org/C190283241 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5141356587409973 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q14599311 |
| concepts[5].display_name | Apoptosis |
| concepts[6].id | https://openalex.org/C53105671 |
| concepts[6].level | 2 |
| concepts[6].score | 0.5077808499336243 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q14777816 |
| concepts[6].display_name | Rational design |
| concepts[7].id | https://openalex.org/C121608353 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4902239441871643 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[7].display_name | Cancer |
| concepts[8].id | https://openalex.org/C96232424 |
| concepts[8].level | 3 |
| concepts[8].score | 0.48250246047973633 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q4118072 |
| concepts[8].display_name | Cancer cell |
| concepts[9].id | https://openalex.org/C154945302 |
| concepts[9].level | 1 |
| concepts[9].score | 0.4653307795524597 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[9].display_name | Artificial intelligence |
| concepts[10].id | https://openalex.org/C70721500 |
| concepts[10].level | 1 |
| concepts[10].score | 0.41513895988464355 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[10].display_name | Computational biology |
| concepts[11].id | https://openalex.org/C60644358 |
| concepts[11].level | 1 |
| concepts[11].score | 0.36928242444992065 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[11].display_name | Bioinformatics |
| concepts[12].id | https://openalex.org/C2776694085 |
| concepts[12].level | 2 |
| concepts[12].score | 0.3437652289867401 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q974135 |
| concepts[12].display_name | Chemotherapy |
| concepts[13].id | https://openalex.org/C86803240 |
| concepts[13].level | 0 |
| concepts[13].score | 0.2743698060512543 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[13].display_name | Biology |
| concepts[14].id | https://openalex.org/C71924100 |
| concepts[14].level | 0 |
| concepts[14].score | 0.22862306237220764 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[14].display_name | Medicine |
| concepts[15].id | https://openalex.org/C171250308 |
| concepts[15].level | 1 |
| concepts[15].score | 0.1858486533164978 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q11468 |
| concepts[15].display_name | Nanotechnology |
| concepts[16].id | https://openalex.org/C126322002 |
| concepts[16].level | 1 |
| concepts[16].score | 0.09440737962722778 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[16].display_name | Internal medicine |
| concepts[17].id | https://openalex.org/C192562407 |
| concepts[17].level | 0 |
| concepts[17].score | 0.09381061792373657 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q228736 |
| concepts[17].display_name | Materials science |
| concepts[18].id | https://openalex.org/C55493867 |
| concepts[18].level | 1 |
| concepts[18].score | 0.0729411244392395 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[18].display_name | Biochemistry |
| concepts[19].id | https://openalex.org/C199360897 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[19].display_name | Programming language |
| keywords[0].id | https://openalex.org/keywords/cisplatin |
| keywords[0].score | 0.7992560267448425 |
| keywords[0].display_name | Cisplatin |
| keywords[1].id | https://openalex.org/keywords/machine-learning |
| keywords[1].score | 0.5869607925415039 |
| keywords[1].display_name | Machine learning |
| keywords[2].id | https://openalex.org/keywords/programmed-cell-death |
| keywords[2].score | 0.5525416135787964 |
| keywords[2].display_name | Programmed cell death |
| keywords[3].id | https://openalex.org/keywords/computer-science |
| keywords[3].score | 0.5395189523696899 |
| keywords[3].display_name | Computer science |
| keywords[4].id | https://openalex.org/keywords/pipeline |
| keywords[4].score | 0.5271360874176025 |
| keywords[4].display_name | Pipeline (software) |
| keywords[5].id | https://openalex.org/keywords/apoptosis |
| keywords[5].score | 0.5141356587409973 |
| keywords[5].display_name | Apoptosis |
| keywords[6].id | https://openalex.org/keywords/rational-design |
| keywords[6].score | 0.5077808499336243 |
| keywords[6].display_name | Rational design |
| keywords[7].id | https://openalex.org/keywords/cancer |
| keywords[7].score | 0.4902239441871643 |
| keywords[7].display_name | Cancer |
| keywords[8].id | https://openalex.org/keywords/cancer-cell |
| keywords[8].score | 0.48250246047973633 |
| keywords[8].display_name | Cancer cell |
| keywords[9].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[9].score | 0.4653307795524597 |
| keywords[9].display_name | Artificial intelligence |
| keywords[10].id | https://openalex.org/keywords/computational-biology |
| keywords[10].score | 0.41513895988464355 |
| keywords[10].display_name | Computational biology |
| keywords[11].id | https://openalex.org/keywords/bioinformatics |
| keywords[11].score | 0.36928242444992065 |
| keywords[11].display_name | Bioinformatics |
| keywords[12].id | https://openalex.org/keywords/chemotherapy |
| keywords[12].score | 0.3437652289867401 |
| keywords[12].display_name | Chemotherapy |
| keywords[13].id | https://openalex.org/keywords/biology |
| keywords[13].score | 0.2743698060512543 |
| keywords[13].display_name | Biology |
| keywords[14].id | https://openalex.org/keywords/medicine |
| keywords[14].score | 0.22862306237220764 |
| keywords[14].display_name | Medicine |
| keywords[15].id | https://openalex.org/keywords/nanotechnology |
| keywords[15].score | 0.1858486533164978 |
| keywords[15].display_name | Nanotechnology |
| keywords[16].id | https://openalex.org/keywords/internal-medicine |
| keywords[16].score | 0.09440737962722778 |
| keywords[16].display_name | Internal medicine |
| keywords[17].id | https://openalex.org/keywords/materials-science |
| keywords[17].score | 0.09381061792373657 |
| keywords[17].display_name | Materials science |
| keywords[18].id | https://openalex.org/keywords/biochemistry |
| keywords[18].score | 0.0729411244392395 |
| keywords[18].display_name | Biochemistry |
| language | en |
| locations[0].id | doi:10.1371/journal.pcbi.1007158 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S86033158 |
| locations[0].source.issn | 1553-734X, 1553-7358 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1553-734X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | PLoS Computational Biology |
| locations[0].source.host_organization | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_name | Public Library of Science |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310315706 |
| locations[0].source.host_organization_lineage_names | Public Library of Science |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printable |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | PLOS Computational Biology |
| locations[0].landing_page_url | https://doi.org/10.1371/journal.pcbi.1007158 |
| locations[1].id | pmid:31498788 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | PLoS computational biology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/31498788 |
| locations[2].id | pmh:oai:repository.arizona.edu:10150/634972 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306400271 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | UA Campus Repository (The University of Arizona) |
| locations[2].source.host_organization | https://openalex.org/I138006243 |
| locations[2].source.host_organization_name | University of Arizona |
| locations[2].source.host_organization_lineage | https://openalex.org/I138006243 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | acceptedVersion |
| locations[2].raw_type | Article |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | True |
| locations[2].is_published | False |
| locations[2].raw_source_name | PLoS computational biology |
| locations[2].landing_page_url | http://hdl.handle.net/10150/634972 |
| locations[3].id | pmh:oai:RePEc:plo:pcbi00:1007158 |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401271 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | RePEc: Research Papers in Economics |
| locations[3].source.host_organization | https://openalex.org/I77793887 |
| locations[3].source.host_organization_name | Federal Reserve Bank of St. Louis |
| locations[3].source.host_organization_lineage | https://openalex.org/I77793887 |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007158 |
| locations[4].id | pmh:oai:doaj.org/article:f501b1749c324b698edad8e4e09907c9 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S4306401280 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[4].source.host_organization | |
| locations[4].source.host_organization_name | |
| locations[4].license | cc-by-sa |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | article |
| locations[4].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | PLoS Computational Biology, Vol 15, Iss 9, p e1007158 (2019) |
| locations[4].landing_page_url | https://doaj.org/article/f501b1749c324b698edad8e4e09907c9 |
| locations[5].id | pmh:oai:europepmc.org:5712052 |
| locations[5].is_oa | True |
| locations[5].source.id | https://openalex.org/S4306400806 |
| locations[5].source.issn | |
| locations[5].source.type | repository |
| locations[5].source.is_oa | False |
| locations[5].source.issn_l | |
| locations[5].source.is_core | False |
| locations[5].source.is_in_doaj | False |
| locations[5].source.display_name | Europe PMC (PubMed Central) |
| locations[5].source.host_organization | https://openalex.org/I1303153112 |
| locations[5].source.host_organization_name | European Bioinformatics Institute |
| locations[5].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[5].license | cc-by |
| locations[5].pdf_url | |
| locations[5].version | submittedVersion |
| locations[5].raw_type | Text |
| locations[5].license_id | https://openalex.org/licenses/cc-by |
| locations[5].is_accepted | False |
| locations[5].is_published | False |
| locations[5].raw_source_name | |
| locations[5].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/6733436 |
| locations[6].id | pmh:oai:figshare.com:article/9786392 |
| locations[6].is_oa | True |
| locations[6].source.id | https://openalex.org/S4306400572 |
| locations[6].source.issn | |
| locations[6].source.type | repository |
| locations[6].source.is_oa | False |
| locations[6].source.issn_l | |
| locations[6].source.is_core | False |
| locations[6].source.is_in_doaj | False |
| locations[6].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[6].source.host_organization | https://openalex.org/I196829312 |
| locations[6].source.host_organization_name | La Trobe University |
| locations[6].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[6].license | cc-by |
| locations[6].pdf_url | |
| locations[6].version | submittedVersion |
| locations[6].raw_type | Dataset |
| locations[6].license_id | https://openalex.org/licenses/cc-by |
| locations[6].is_accepted | False |
| locations[6].is_published | False |
| locations[6].raw_source_name | |
| locations[6].landing_page_url | https://figshare.com/articles/dataset/Designing_combination_therapies_with_modeling_chaperoned_machine_learning/9786392 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5002416203 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-6127-7535 |
| authorships[0].author.display_name | Yin Zhang |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I1285204247 |
| authorships[0].affiliations[0].raw_affiliation_string | Division of Biostatistics and Epidemiology, Cincinnati Children's Hospital Medical Center, Cincinnati, Ohio, United States of America |
| authorships[0].institutions[0].id | https://openalex.org/I1285204247 |
| authorships[0].institutions[0].ror | https://ror.org/01hcyya48 |
| authorships[0].institutions[0].type | healthcare |
| authorships[0].institutions[0].lineage | https://openalex.org/I1285204247 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Cincinnati Children's Hospital Medical Center |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Yin Zhang |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | Division of Biostatistics and Epidemiology, Cincinnati Children's Hospital Medical Center, Cincinnati, Ohio, United States of America |
| authorships[1].author.id | https://openalex.org/A5081333468 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Julie M. Huynh |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I138006243 |
| authorships[1].affiliations[0].raw_affiliation_string | Molecular and Cellular Biology, University of Arizona, Tucson, United States of America |
| authorships[1].institutions[0].id | https://openalex.org/I138006243 |
| authorships[1].institutions[0].ror | https://ror.org/03m2x1q45 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I138006243 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | University of Arizona |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Julie M. Huynh |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Molecular and Cellular Biology, University of Arizona, Tucson, United States of America |
| authorships[2].author.id | https://openalex.org/A5101422397 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-3255-2777 |
| authorships[2].author.display_name | Guan‐Sheng Liu |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I63135867 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Pharmacology and Systems Physiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, United States of America |
| authorships[2].institutions[0].id | https://openalex.org/I63135867 |
| authorships[2].institutions[0].ror | https://ror.org/01e3m7079 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I63135867 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | University of Cincinnati |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Guan-Sheng Liu |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | Department of Pharmacology and Systems Physiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, United States of America |
| authorships[3].author.id | https://openalex.org/A5014243011 |
| authorships[3].author.orcid | https://orcid.org/0000-0001-7872-2941 |
| authorships[3].author.display_name | Richard Ballweg |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I63135867 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Pharmacology and Systems Physiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, United States of America |
| authorships[3].institutions[0].id | https://openalex.org/I63135867 |
| authorships[3].institutions[0].ror | https://ror.org/01e3m7079 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I63135867 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of Cincinnati |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Richard Ballweg |
| authorships[3].is_corresponding | True |
| authorships[3].raw_affiliation_strings | Department of Pharmacology and Systems Physiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, United States of America |
| authorships[4].author.id | https://openalex.org/A5050015576 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Kayenat S. Aryeh |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I138006243 |
| authorships[4].affiliations[0].raw_affiliation_string | Molecular and Cellular Biology, University of Arizona, Tucson, United States of America |
| authorships[4].institutions[0].id | https://openalex.org/I138006243 |
| authorships[4].institutions[0].ror | https://ror.org/03m2x1q45 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I138006243 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | University of Arizona |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Kayenat S. Aryeh |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | Molecular and Cellular Biology, University of Arizona, Tucson, United States of America |
| authorships[5].author.id | https://openalex.org/A5054257493 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-2835-8544 |
| authorships[5].author.display_name | Andrew L. Paek |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I138006243 |
| authorships[5].affiliations[0].raw_affiliation_string | Molecular and Cellular Biology, University of Arizona, Tucson, United States of America |
| authorships[5].institutions[0].id | https://openalex.org/I138006243 |
| authorships[5].institutions[0].ror | https://ror.org/03m2x1q45 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I138006243 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | University of Arizona |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Andrew L. Paek |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Molecular and Cellular Biology, University of Arizona, Tucson, United States of America |
| authorships[6].author.id | https://openalex.org/A5003112405 |
| authorships[6].author.orcid | https://orcid.org/0000-0003-1773-6279 |
| authorships[6].author.display_name | Tongli Zhang |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I63135867 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Pharmacology and Systems Physiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, United States of America |
| authorships[6].institutions[0].id | https://openalex.org/I63135867 |
| authorships[6].institutions[0].ror | https://ror.org/01e3m7079 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I63135867 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | University of Cincinnati |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Tongli Zhang |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Department of Pharmacology and Systems Physiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, United States of America |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printable |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Designing combination therapies with modeling chaperoned machine learning |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10211 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9994999766349792 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1703 |
| primary_topic.subfield.display_name | Computational Theory and Mathematics |
| primary_topic.display_name | Computational Drug Discovery Methods |
| related_works | https://openalex.org/W2091461259, https://openalex.org/W2149068418, https://openalex.org/W2406608798, https://openalex.org/W2406596433, https://openalex.org/W2080420195, https://openalex.org/W2066496973, https://openalex.org/W2460036924, https://openalex.org/W2021865845, https://openalex.org/W3120025342, https://openalex.org/W1971929983 |
| cited_by_count | 15 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 1 |
| counts_by_year[3].year | 2022 |
| counts_by_year[3].cited_by_count | 2 |
| counts_by_year[4].year | 2021 |
| counts_by_year[4].cited_by_count | 4 |
| counts_by_year[5].year | 2020 |
| counts_by_year[5].cited_by_count | 3 |
| locations_count | 7 |
| best_oa_location.id | doi:10.1371/journal.pcbi.1007158 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S86033158 |
| best_oa_location.source.issn | 1553-734X, 1553-7358 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1553-734X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | PLoS Computational Biology |
| best_oa_location.source.host_organization | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_name | Public Library of Science |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| best_oa_location.source.host_organization_lineage_names | Public Library of Science |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printable |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | PLOS Computational Biology |
| best_oa_location.landing_page_url | https://doi.org/10.1371/journal.pcbi.1007158 |
| primary_location.id | doi:10.1371/journal.pcbi.1007158 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S86033158 |
| primary_location.source.issn | 1553-734X, 1553-7358 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1553-734X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | PLoS Computational Biology |
| primary_location.source.host_organization | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_name | Public Library of Science |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310315706 |
| primary_location.source.host_organization_lineage_names | Public Library of Science |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1007158&type=printable |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | PLOS Computational Biology |
| primary_location.landing_page_url | https://doi.org/10.1371/journal.pcbi.1007158 |
| publication_date | 2019-09-09 |
| publication_year | 2019 |
| referenced_works | https://openalex.org/W2131487381, https://openalex.org/W2604956011, https://openalex.org/W2102382046, https://openalex.org/W2042619042, https://openalex.org/W2153612800, https://openalex.org/W2296577107, https://openalex.org/W2045001281, https://openalex.org/W2132672330, https://openalex.org/W2111510713, https://openalex.org/W2321162622, https://openalex.org/W2313997600, https://openalex.org/W2023168478, https://openalex.org/W1925794468, https://openalex.org/W2795843903, https://openalex.org/W2903992755, https://openalex.org/W2787894218, https://openalex.org/W2169608095, https://openalex.org/W2158398965, https://openalex.org/W2059340786, https://openalex.org/W2135777283, https://openalex.org/W2092176773, https://openalex.org/W2173690212, https://openalex.org/W2081116284, https://openalex.org/W2033671436, https://openalex.org/W1991083334, https://openalex.org/W2197234859, https://openalex.org/W2150550712, https://openalex.org/W2094550799, https://openalex.org/W2144370915, https://openalex.org/W2116944463, https://openalex.org/W2142059159, https://openalex.org/W2139183953, https://openalex.org/W2774112395, https://openalex.org/W2001576594, https://openalex.org/W2163482104, https://openalex.org/W2805310212, https://openalex.org/W2951934944, https://openalex.org/W2790102940, https://openalex.org/W2804500674, https://openalex.org/W2951315938, https://openalex.org/W2908302506, https://openalex.org/W2904637438, https://openalex.org/W2141997480, https://openalex.org/W2097651998, https://openalex.org/W1995354282, https://openalex.org/W2743509530, https://openalex.org/W1554944419 |
| referenced_works_count | 47 |
| abstract_inverted_index.a | 3, 13, 40, 77, 91, 120, 137 |
| abstract_inverted_index.In | 29 |
| abstract_inverted_index.an | 124, 173 |
| abstract_inverted_index.be | 194 |
| abstract_inverted_index.by | 98, 136 |
| abstract_inverted_index.in | 110 |
| abstract_inverted_index.is | 2, 134 |
| abstract_inverted_index.of | 10, 20, 35, 58, 80, 102, 126, 172, 186, 205 |
| abstract_inverted_index.on | 54 |
| abstract_inverted_index.or | 64 |
| abstract_inverted_index.to | 6, 31, 62, 119, 151, 196 |
| abstract_inverted_index.we | 38, 165 |
| abstract_inverted_index.The | 73, 113, 155 |
| abstract_inverted_index.and | 50, 82, 89, 107 |
| abstract_inverted_index.are | 158 |
| abstract_inverted_index.can | 147, 178, 192 |
| abstract_inverted_index.for | 16 |
| abstract_inverted_index.our | 180 |
| abstract_inverted_index.p53 | 103 |
| abstract_inverted_index.the | 7, 17, 46, 55, 68, 100, 184, 203 |
| abstract_inverted_index.was | 96, 116 |
| abstract_inverted_index.This | 190 |
| abstract_inverted_index.cell | 87, 105, 108 |
| abstract_inverted_index.data | 53 |
| abstract_inverted_index.each | 132 |
| abstract_inverted_index.have | 166 |
| abstract_inverted_index.huge | 26 |
| abstract_inverted_index.into | 183 |
| abstract_inverted_index.that | 44, 76, 146, 168, 200 |
| abstract_inverted_index.then | 117, 193 |
| abstract_inverted_index.they | 140 |
| abstract_inverted_index.with | 67, 123, 160 |
| abstract_inverted_index.TRAIL | 83 |
| abstract_inverted_index.Thus, | 12 |
| abstract_inverted_index.bring | 25 |
| abstract_inverted_index.cells | 61, 150 |
| abstract_inverted_index.could | 24, 201 |
| abstract_inverted_index.death | 88, 109 |
| abstract_inverted_index.fate, | 106 |
| abstract_inverted_index.major | 4 |
| abstract_inverted_index.model | 43, 115, 177 |
| abstract_inverted_index.order | 30 |
| abstract_inverted_index.tumor | 149 |
| abstract_inverted_index.would | 84 |
| abstract_inverted_index.Though | 131 |
| abstract_inverted_index.cancer | 60, 86 |
| abstract_inverted_index.cells. | 112 |
| abstract_inverted_index.design | 34, 197 |
| abstract_inverted_index.ligand | 71 |
| abstract_inverted_index.method | 133 |
| abstract_inverted_index.single | 111 |
| abstract_inverted_index.cancer. | 11 |
| abstract_inverted_index.convert | 179 |
| abstract_inverted_index.diverse | 127 |
| abstract_inverted_index.enhance | 85 |
| abstract_inverted_index.exhibit | 90 |
| abstract_inverted_index.improve | 202 |
| abstract_inverted_index.machine | 128, 169 |
| abstract_inverted_index.model's | 74 |
| abstract_inverted_index.players | 145 |
| abstract_inverted_index.several | 143 |
| abstract_inverted_index.(TRAIL). | 72 |
| abstract_inverted_index.Overall, | 164 |
| abstract_inverted_index.analysis | 122, 171 |
| abstract_inverted_index.benefit. | 28 |
| abstract_inverted_index.combined | 66 |
| abstract_inverted_index.dynamics | 101 |
| abstract_inverted_index.efficacy | 204 |
| abstract_inverted_index.ensemble | 125 |
| abstract_inverted_index.identity | 185 |
| abstract_inverted_index.killing" | 93 |
| abstract_inverted_index.learning | 129, 170 |
| abstract_inverted_index.methods. | 130 |
| abstract_inverted_index.pattern, | 95 |
| abstract_inverted_index.pipeline | 15 |
| abstract_inverted_index.previous | 161 |
| abstract_inverted_index.rational | 33 |
| abstract_inverted_index.relevant | 51 |
| abstract_inverted_index.response | 57 |
| abstract_inverted_index.temporal | 94 |
| abstract_inverted_index."two-wave | 92 |
| abstract_inverted_index.apoptosis | 153 |
| abstract_inverted_index.available | 47, 181 |
| abstract_inverted_index.challenge | 5 |
| abstract_inverted_index.cisplatin | 63, 65, 81 |
| abstract_inverted_index.developed | 39 |
| abstract_inverted_index.different | 138 |
| abstract_inverted_index.effective | 8, 21 |
| abstract_inverted_index.efficient | 18 |
| abstract_inverted_index.knowledge | 49, 182, 191 |
| abstract_inverted_index.leveraged | 195 |
| abstract_inverted_index.measuring | 99 |
| abstract_inverted_index.molecular | 144 |
| abstract_inverted_index.sensitize | 148 |
| abstract_inverted_index.subjected | 118 |
| abstract_inverted_index.treatment | 9, 79, 198 |
| abstract_inverted_index.validated | 97, 114, 175 |
| abstract_inverted_index.algorithm, | 139 |
| abstract_inverted_index.biological | 48 |
| abstract_inverted_index.biomedical | 27 |
| abstract_inverted_index.consistent | 159 |
| abstract_inverted_index.facilitate | 32 |
| abstract_inverted_index.identified | 142, 156 |
| abstract_inverted_index.individual | 59 |
| abstract_inverted_index.meaningful | 188 |
| abstract_inverted_index.resistance | 1 |
| abstract_inverted_index.strategies | 199 |
| abstract_inverted_index.systematic | 14, 121 |
| abstract_inverted_index.therapies, | 37 |
| abstract_inverted_index.treatments | 23 |
| abstract_inverted_index.TNF-related | 69 |
| abstract_inverted_index.combination | 22, 36, 78 |
| abstract_inverted_index.illustrated | 167 |
| abstract_inverted_index.mechanistic | 176 |
| abstract_inverted_index.sensitizers | 157 |
| abstract_inverted_index.Chemotherapy | 0 |
| abstract_inverted_index.biologically | 187 |
| abstract_inverted_index.collectively | 141 |
| abstract_inverted_index.experimental | 52, 162 |
| abstract_inverted_index.incorporates | 45 |
| abstract_inverted_index.predictions, | 75 |
| abstract_inverted_index.sensitizers. | 189 |
| abstract_inverted_index.accumulation, | 104 |
| abstract_inverted_index.characterized | 135 |
| abstract_inverted_index.chemotherapy. | 206 |
| abstract_inverted_index.comprehensive | 41 |
| abstract_inverted_index.computational | 42 |
| abstract_inverted_index.observations. | 163 |
| abstract_inverted_index.(sensitizers). | 154 |
| abstract_inverted_index.experimentally | 174 |
| abstract_inverted_index.identification | 19 |
| abstract_inverted_index.life-and-death | 56 |
| abstract_inverted_index.cisplatin-induced | 152 |
| abstract_inverted_index.apoptosis-inducing | 70 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5101422397, https://openalex.org/A5003112405, https://openalex.org/A5081333468, https://openalex.org/A5014243011, https://openalex.org/A5002416203, https://openalex.org/A5054257493, https://openalex.org/A5050015576 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I1285204247, https://openalex.org/I138006243, https://openalex.org/I63135867 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.7900000214576721 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.87131474 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |