Development and Validation of Fully Automatic Deep Learning-Based Algorithms for Immunohistochemistry Reporting of Invasive Breast Ductal Carcinoma Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.48550/arxiv.2406.10893
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.48550/arxiv.2406.10893
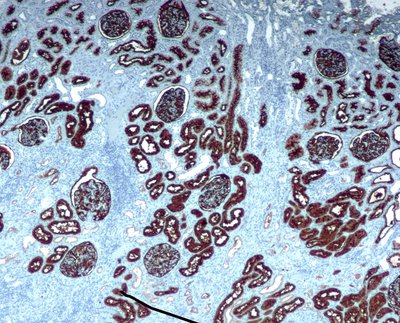
Immunohistochemistry (IHC) analysis is a well-accepted and widely used method for molecular subtyping, a procedure for prognosis and targeted therapy of breast carcinoma, the most common type of tumor affecting women. There are four molecular biomarkers namely progesterone receptor (PR), estrogen receptor (ER), antigen Ki67, and human epidermal growth factor receptor 2 (HER2) whose assessment is needed under IHC procedure to decide prognosis as well as predictors of response to therapy. However, IHC scoring is based on subjective microscopic examination of tumor morphology and suffers from poor reproducibility, high subjectivity, and often incorrect scoring in low-score cases. In this paper, we present, a deep learning-based semi-supervised trained, fully automatic, decision support system (DSS) for IHC scoring of invasive ductal carcinoma. Our system automatically detects the tumor region removing artifacts and scores based on Allred standard. The system is developed using 3 million pathologist-annotated image patches from 300 slides, fifty thousand in-house cell annotations, and forty thousand pixels marking of HER2 membrane. We have conducted multicentric trials at four centers with three different types of digital scanners in terms of percentage agreement with doctors. And achieved agreements of 95, 92, 88 and 82 percent for Ki67, HER2, ER, and PR stain categories, respectively. In addition to overall accuracy, we found that there is 5 percent of cases where pathologist have changed their score in favor of algorithm score while reviewing with detailed algorithmic analysis. Our approach could improve the accuracy of IHC scoring and subsequent therapy decisions, particularly where specialist expertise is unavailable. Our system is highly modular. The proposed algorithm modules can be used to develop DSS for other cancer types.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- http://arxiv.org/abs/2406.10893
- https://arxiv.org/pdf/2406.10893
- OA Status
- green
- Cited By
- 1
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4399794647
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4399794647Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.48550/arxiv.2406.10893Digital Object Identifier
- Title
-
Development and Validation of Fully Automatic Deep Learning-Based Algorithms for Immunohistochemistry Reporting of Invasive Breast Ductal CarcinomaWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-06-16Full publication date if available
- Authors
-
Sumit Kumar Jha, Purnendu Mishra, Shubham Mathur, Gursewak Singh, Rajiv Kumar, Kiran Aatre, Suraj RengarajanList of authors in order
- Landing page
-
https://arxiv.org/abs/2406.10893Publisher landing page
- PDF URL
-
https://arxiv.org/pdf/2406.10893Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://arxiv.org/pdf/2406.10893Direct OA link when available
- Concepts
-
Immunohistochemistry, Ductal carcinoma, Algorithm, Breast carcinoma, Computer science, Carcinoma, Artificial intelligence, Medicine, Pathology, Breast cancer, Internal medicine, CancerTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2024: 1Per-year citation counts (last 5 years)
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4399794647 |
|---|---|
| doi | https://doi.org/10.48550/arxiv.2406.10893 |
| ids.doi | https://doi.org/10.48550/arxiv.2406.10893 |
| ids.openalex | https://openalex.org/W4399794647 |
| fwci | |
| type | preprint |
| title | Development and Validation of Fully Automatic Deep Learning-Based Algorithms for Immunohistochemistry Reporting of Invasive Breast Ductal Carcinoma |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10862 |
| topics[0].field.id | https://openalex.org/fields/17 |
| topics[0].field.display_name | Computer Science |
| topics[0].score | 0.9944999814033508 |
| topics[0].domain.id | https://openalex.org/domains/3 |
| topics[0].domain.display_name | Physical Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1702 |
| topics[0].subfield.display_name | Artificial Intelligence |
| topics[0].display_name | AI in cancer detection |
| topics[1].id | https://openalex.org/T12422 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9609000086784363 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2741 |
| topics[1].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[1].display_name | Radiomics and Machine Learning in Medical Imaging |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C204232928 |
| concepts[0].level | 2 |
| concepts[0].score | 0.6326323747634888 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q899285 |
| concepts[0].display_name | Immunohistochemistry |
| concepts[1].id | https://openalex.org/C2780862961 |
| concepts[1].level | 4 |
| concepts[1].score | 0.49214857816696167 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q5311598 |
| concepts[1].display_name | Ductal carcinoma |
| concepts[2].id | https://openalex.org/C11413529 |
| concepts[2].level | 1 |
| concepts[2].score | 0.47455650568008423 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q8366 |
| concepts[2].display_name | Algorithm |
| concepts[3].id | https://openalex.org/C3018521938 |
| concepts[3].level | 4 |
| concepts[3].score | 0.4399912357330322 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q128581 |
| concepts[3].display_name | Breast carcinoma |
| concepts[4].id | https://openalex.org/C41008148 |
| concepts[4].level | 0 |
| concepts[4].score | 0.4348428249359131 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[4].display_name | Computer science |
| concepts[5].id | https://openalex.org/C2777546739 |
| concepts[5].level | 2 |
| concepts[5].score | 0.4280340075492859 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q33525 |
| concepts[5].display_name | Carcinoma |
| concepts[6].id | https://openalex.org/C154945302 |
| concepts[6].level | 1 |
| concepts[6].score | 0.4249691367149353 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[6].display_name | Artificial intelligence |
| concepts[7].id | https://openalex.org/C71924100 |
| concepts[7].level | 0 |
| concepts[7].score | 0.2940724194049835 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[7].display_name | Medicine |
| concepts[8].id | https://openalex.org/C142724271 |
| concepts[8].level | 1 |
| concepts[8].score | 0.28249651193618774 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[8].display_name | Pathology |
| concepts[9].id | https://openalex.org/C530470458 |
| concepts[9].level | 3 |
| concepts[9].score | 0.26505720615386963 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q128581 |
| concepts[9].display_name | Breast cancer |
| concepts[10].id | https://openalex.org/C126322002 |
| concepts[10].level | 1 |
| concepts[10].score | 0.2025623917579651 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[10].display_name | Internal medicine |
| concepts[11].id | https://openalex.org/C121608353 |
| concepts[11].level | 2 |
| concepts[11].score | 0.11402150988578796 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[11].display_name | Cancer |
| keywords[0].id | https://openalex.org/keywords/immunohistochemistry |
| keywords[0].score | 0.6326323747634888 |
| keywords[0].display_name | Immunohistochemistry |
| keywords[1].id | https://openalex.org/keywords/ductal-carcinoma |
| keywords[1].score | 0.49214857816696167 |
| keywords[1].display_name | Ductal carcinoma |
| keywords[2].id | https://openalex.org/keywords/algorithm |
| keywords[2].score | 0.47455650568008423 |
| keywords[2].display_name | Algorithm |
| keywords[3].id | https://openalex.org/keywords/breast-carcinoma |
| keywords[3].score | 0.4399912357330322 |
| keywords[3].display_name | Breast carcinoma |
| keywords[4].id | https://openalex.org/keywords/computer-science |
| keywords[4].score | 0.4348428249359131 |
| keywords[4].display_name | Computer science |
| keywords[5].id | https://openalex.org/keywords/carcinoma |
| keywords[5].score | 0.4280340075492859 |
| keywords[5].display_name | Carcinoma |
| keywords[6].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[6].score | 0.4249691367149353 |
| keywords[6].display_name | Artificial intelligence |
| keywords[7].id | https://openalex.org/keywords/medicine |
| keywords[7].score | 0.2940724194049835 |
| keywords[7].display_name | Medicine |
| keywords[8].id | https://openalex.org/keywords/pathology |
| keywords[8].score | 0.28249651193618774 |
| keywords[8].display_name | Pathology |
| keywords[9].id | https://openalex.org/keywords/breast-cancer |
| keywords[9].score | 0.26505720615386963 |
| keywords[9].display_name | Breast cancer |
| keywords[10].id | https://openalex.org/keywords/internal-medicine |
| keywords[10].score | 0.2025623917579651 |
| keywords[10].display_name | Internal medicine |
| keywords[11].id | https://openalex.org/keywords/cancer |
| keywords[11].score | 0.11402150988578796 |
| keywords[11].display_name | Cancer |
| language | en |
| locations[0].id | pmh:oai:arXiv.org:2406.10893 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400194 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | arXiv (Cornell University) |
| locations[0].source.host_organization | https://openalex.org/I205783295 |
| locations[0].source.host_organization_name | Cornell University |
| locations[0].source.host_organization_lineage | https://openalex.org/I205783295 |
| locations[0].license | |
| locations[0].pdf_url | https://arxiv.org/pdf/2406.10893 |
| locations[0].version | submittedVersion |
| locations[0].raw_type | |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | http://arxiv.org/abs/2406.10893 |
| locations[1].id | doi:10.48550/arxiv.2406.10893 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400194 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | True |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | arXiv (Cornell University) |
| locations[1].source.host_organization | https://openalex.org/I205783295 |
| locations[1].source.host_organization_name | Cornell University |
| locations[1].source.host_organization_lineage | https://openalex.org/I205783295 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | |
| locations[1].raw_type | article |
| locations[1].license_id | |
| locations[1].is_accepted | False |
| locations[1].is_published | |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://doi.org/10.48550/arxiv.2406.10893 |
| indexed_in | arxiv, datacite |
| authorships[0].author.id | https://openalex.org/A5075978538 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-0354-2940 |
| authorships[0].author.display_name | Sumit Kumar Jha |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jha, Sumit Kumar |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5032671035 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-2060-8972 |
| authorships[1].author.display_name | Purnendu Mishra |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mishra, Purnendu |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5012464938 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-5616-0877 |
| authorships[2].author.display_name | Shubham Mathur |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Mathur, Shubham |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5102300081 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Gursewak Singh |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Singh, Gursewak |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5002969277 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-7774-2673 |
| authorships[4].author.display_name | Rajiv Kumar |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Kumar, Rajiv |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5073519483 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Kiran Aatre |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Aatre, Kiran |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5104330981 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Suraj Rengarajan |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Rengarajan, Suraj |
| authorships[6].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://arxiv.org/pdf/2406.10893 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2024-06-19T00:00:00 |
| display_name | Development and Validation of Fully Automatic Deep Learning-Based Algorithms for Immunohistochemistry Reporting of Invasive Breast Ductal Carcinoma |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T10862 |
| primary_topic.field.id | https://openalex.org/fields/17 |
| primary_topic.field.display_name | Computer Science |
| primary_topic.score | 0.9944999814033508 |
| primary_topic.domain.id | https://openalex.org/domains/3 |
| primary_topic.domain.display_name | Physical Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1702 |
| primary_topic.subfield.display_name | Artificial Intelligence |
| primary_topic.display_name | AI in cancer detection |
| related_works | https://openalex.org/W2051487156, https://openalex.org/W2999877485, https://openalex.org/W2073681303, https://openalex.org/W2355920994, https://openalex.org/W2399421402, https://openalex.org/W2373084812, https://openalex.org/W2464212128, https://openalex.org/W2113335236, https://openalex.org/W2328395017, https://openalex.org/W2410574247 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2024 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 2 |
| best_oa_location.id | pmh:oai:arXiv.org:2406.10893 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400194 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | arXiv (Cornell University) |
| best_oa_location.source.host_organization | https://openalex.org/I205783295 |
| best_oa_location.source.host_organization_name | Cornell University |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I205783295 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://arxiv.org/pdf/2406.10893 |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | http://arxiv.org/abs/2406.10893 |
| primary_location.id | pmh:oai:arXiv.org:2406.10893 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400194 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | arXiv (Cornell University) |
| primary_location.source.host_organization | https://openalex.org/I205783295 |
| primary_location.source.host_organization_name | Cornell University |
| primary_location.source.host_organization_lineage | https://openalex.org/I205783295 |
| primary_location.license | |
| primary_location.pdf_url | https://arxiv.org/pdf/2406.10893 |
| primary_location.version | submittedVersion |
| primary_location.raw_type | |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | http://arxiv.org/abs/2406.10893 |
| publication_date | 2024-06-16 |
| publication_year | 2024 |
| referenced_works_count | 0 |
| abstract_inverted_index.2 | 51 |
| abstract_inverted_index.3 | 140 |
| abstract_inverted_index.5 | 212 |
| abstract_inverted_index.a | 4, 13, 102 |
| abstract_inverted_index.82 | 191 |
| abstract_inverted_index.88 | 189 |
| abstract_inverted_index.In | 97, 202 |
| abstract_inverted_index.PR | 198 |
| abstract_inverted_index.We | 161 |
| abstract_inverted_index.as | 63, 65 |
| abstract_inverted_index.at | 166 |
| abstract_inverted_index.be | 262 |
| abstract_inverted_index.in | 94, 176, 222 |
| abstract_inverted_index.is | 3, 55, 74, 137, 211, 250, 254 |
| abstract_inverted_index.of | 20, 27, 67, 80, 116, 158, 173, 178, 186, 214, 224, 239 |
| abstract_inverted_index.on | 76, 132 |
| abstract_inverted_index.to | 60, 69, 204, 264 |
| abstract_inverted_index.we | 100, 207 |
| abstract_inverted_index.300 | 146 |
| abstract_inverted_index.92, | 188 |
| abstract_inverted_index.95, | 187 |
| abstract_inverted_index.And | 183 |
| abstract_inverted_index.DSS | 266 |
| abstract_inverted_index.ER, | 196 |
| abstract_inverted_index.IHC | 58, 72, 114, 240 |
| abstract_inverted_index.Our | 120, 233, 252 |
| abstract_inverted_index.The | 135, 257 |
| abstract_inverted_index.and | 6, 17, 45, 83, 90, 129, 153, 190, 197, 242 |
| abstract_inverted_index.are | 32 |
| abstract_inverted_index.can | 261 |
| abstract_inverted_index.for | 10, 15, 113, 193, 267 |
| abstract_inverted_index.the | 23, 124, 237 |
| abstract_inverted_index.HER2 | 159 |
| abstract_inverted_index.cell | 151 |
| abstract_inverted_index.deep | 103 |
| abstract_inverted_index.four | 33, 167 |
| abstract_inverted_index.from | 85, 145 |
| abstract_inverted_index.have | 162, 218 |
| abstract_inverted_index.high | 88 |
| abstract_inverted_index.most | 24 |
| abstract_inverted_index.poor | 86 |
| abstract_inverted_index.that | 209 |
| abstract_inverted_index.this | 98 |
| abstract_inverted_index.type | 26 |
| abstract_inverted_index.used | 8, 263 |
| abstract_inverted_index.well | 64 |
| abstract_inverted_index.with | 169, 181, 229 |
| abstract_inverted_index.(DSS) | 112 |
| abstract_inverted_index.(ER), | 42 |
| abstract_inverted_index.(IHC) | 1 |
| abstract_inverted_index.(PR), | 39 |
| abstract_inverted_index.HER2, | 195 |
| abstract_inverted_index.Ki67, | 44, 194 |
| abstract_inverted_index.There | 31 |
| abstract_inverted_index.based | 75, 131 |
| abstract_inverted_index.cases | 215 |
| abstract_inverted_index.could | 235 |
| abstract_inverted_index.favor | 223 |
| abstract_inverted_index.fifty | 148 |
| abstract_inverted_index.forty | 154 |
| abstract_inverted_index.found | 208 |
| abstract_inverted_index.fully | 107 |
| abstract_inverted_index.human | 46 |
| abstract_inverted_index.image | 143 |
| abstract_inverted_index.often | 91 |
| abstract_inverted_index.other | 268 |
| abstract_inverted_index.score | 221, 226 |
| abstract_inverted_index.stain | 199 |
| abstract_inverted_index.terms | 177 |
| abstract_inverted_index.their | 220 |
| abstract_inverted_index.there | 210 |
| abstract_inverted_index.three | 170 |
| abstract_inverted_index.tumor | 28, 81, 125 |
| abstract_inverted_index.types | 172 |
| abstract_inverted_index.under | 57 |
| abstract_inverted_index.using | 139 |
| abstract_inverted_index.where | 216, 247 |
| abstract_inverted_index.while | 227 |
| abstract_inverted_index.whose | 53 |
| abstract_inverted_index.(HER2) | 52 |
| abstract_inverted_index.Allred | 133 |
| abstract_inverted_index.breast | 21 |
| abstract_inverted_index.cancer | 269 |
| abstract_inverted_index.cases. | 96 |
| abstract_inverted_index.common | 25 |
| abstract_inverted_index.decide | 61 |
| abstract_inverted_index.ductal | 118 |
| abstract_inverted_index.factor | 49 |
| abstract_inverted_index.growth | 48 |
| abstract_inverted_index.highly | 255 |
| abstract_inverted_index.method | 9 |
| abstract_inverted_index.namely | 36 |
| abstract_inverted_index.needed | 56 |
| abstract_inverted_index.paper, | 99 |
| abstract_inverted_index.pixels | 156 |
| abstract_inverted_index.region | 126 |
| abstract_inverted_index.scores | 130 |
| abstract_inverted_index.system | 111, 121, 136, 253 |
| abstract_inverted_index.trials | 165 |
| abstract_inverted_index.types. | 270 |
| abstract_inverted_index.widely | 7 |
| abstract_inverted_index.women. | 30 |
| abstract_inverted_index.antigen | 43 |
| abstract_inverted_index.centers | 168 |
| abstract_inverted_index.changed | 219 |
| abstract_inverted_index.detects | 123 |
| abstract_inverted_index.develop | 265 |
| abstract_inverted_index.digital | 174 |
| abstract_inverted_index.improve | 236 |
| abstract_inverted_index.marking | 157 |
| abstract_inverted_index.million | 141 |
| abstract_inverted_index.modules | 260 |
| abstract_inverted_index.overall | 205 |
| abstract_inverted_index.patches | 144 |
| abstract_inverted_index.percent | 192, 213 |
| abstract_inverted_index.scoring | 73, 93, 115, 241 |
| abstract_inverted_index.slides, | 147 |
| abstract_inverted_index.suffers | 84 |
| abstract_inverted_index.support | 110 |
| abstract_inverted_index.therapy | 19, 244 |
| abstract_inverted_index.However, | 71 |
| abstract_inverted_index.accuracy | 238 |
| abstract_inverted_index.achieved | 184 |
| abstract_inverted_index.addition | 203 |
| abstract_inverted_index.analysis | 2 |
| abstract_inverted_index.approach | 234 |
| abstract_inverted_index.decision | 109 |
| abstract_inverted_index.detailed | 230 |
| abstract_inverted_index.doctors. | 182 |
| abstract_inverted_index.estrogen | 40 |
| abstract_inverted_index.in-house | 150 |
| abstract_inverted_index.invasive | 117 |
| abstract_inverted_index.modular. | 256 |
| abstract_inverted_index.present, | 101 |
| abstract_inverted_index.proposed | 258 |
| abstract_inverted_index.receptor | 38, 41, 50 |
| abstract_inverted_index.removing | 127 |
| abstract_inverted_index.response | 68 |
| abstract_inverted_index.scanners | 175 |
| abstract_inverted_index.targeted | 18 |
| abstract_inverted_index.therapy. | 70 |
| abstract_inverted_index.thousand | 149, 155 |
| abstract_inverted_index.trained, | 106 |
| abstract_inverted_index.accuracy, | 206 |
| abstract_inverted_index.affecting | 29 |
| abstract_inverted_index.agreement | 180 |
| abstract_inverted_index.algorithm | 225, 259 |
| abstract_inverted_index.analysis. | 232 |
| abstract_inverted_index.artifacts | 128 |
| abstract_inverted_index.conducted | 163 |
| abstract_inverted_index.developed | 138 |
| abstract_inverted_index.different | 171 |
| abstract_inverted_index.epidermal | 47 |
| abstract_inverted_index.expertise | 249 |
| abstract_inverted_index.incorrect | 92 |
| abstract_inverted_index.low-score | 95 |
| abstract_inverted_index.membrane. | 160 |
| abstract_inverted_index.molecular | 11, 34 |
| abstract_inverted_index.procedure | 14, 59 |
| abstract_inverted_index.prognosis | 16, 62 |
| abstract_inverted_index.reviewing | 228 |
| abstract_inverted_index.standard. | 134 |
| abstract_inverted_index.agreements | 185 |
| abstract_inverted_index.assessment | 54 |
| abstract_inverted_index.automatic, | 108 |
| abstract_inverted_index.biomarkers | 35 |
| abstract_inverted_index.carcinoma, | 22 |
| abstract_inverted_index.carcinoma. | 119 |
| abstract_inverted_index.decisions, | 245 |
| abstract_inverted_index.morphology | 82 |
| abstract_inverted_index.percentage | 179 |
| abstract_inverted_index.predictors | 66 |
| abstract_inverted_index.specialist | 248 |
| abstract_inverted_index.subjective | 77 |
| abstract_inverted_index.subsequent | 243 |
| abstract_inverted_index.subtyping, | 12 |
| abstract_inverted_index.algorithmic | 231 |
| abstract_inverted_index.categories, | 200 |
| abstract_inverted_index.examination | 79 |
| abstract_inverted_index.microscopic | 78 |
| abstract_inverted_index.pathologist | 217 |
| abstract_inverted_index.annotations, | 152 |
| abstract_inverted_index.multicentric | 164 |
| abstract_inverted_index.particularly | 246 |
| abstract_inverted_index.progesterone | 37 |
| abstract_inverted_index.unavailable. | 251 |
| abstract_inverted_index.automatically | 122 |
| abstract_inverted_index.respectively. | 201 |
| abstract_inverted_index.subjectivity, | 89 |
| abstract_inverted_index.well-accepted | 5 |
| abstract_inverted_index.learning-based | 104 |
| abstract_inverted_index.semi-supervised | 105 |
| abstract_inverted_index.reproducibility, | 87 |
| abstract_inverted_index.Immunohistochemistry | 0 |
| abstract_inverted_index.pathologist-annotated | 142 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 7 |
| citation_normalized_percentile |