Diff-AMP: tailored designed antimicrobial peptide framework with all-in-one generation, identification, prediction and optimization Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1093/bib/bbae078
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1093/bib/bbae078
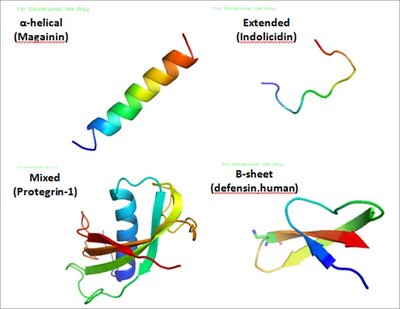
Antimicrobial peptides (AMPs), short peptides with diverse functions, effectively target and combat various organisms. The widespread misuse of chemical antibiotics has led to increasing microbial resistance. Due to their low drug resistance and toxicity, AMPs are considered promising substitutes for traditional antibiotics. While existing deep learning technology enhances AMP generation, it also presents certain challenges. Firstly, AMP generation overlooks the complex interdependencies among amino acids. Secondly, current models fail to integrate crucial tasks like screening, attribute prediction and iterative optimization. Consequently, we develop a integrated deep learning framework, Diff-AMP, that automates AMP generation, identification, attribute prediction and iterative optimization. We innovatively integrate kinetic diffusion and attention mechanisms into the reinforcement learning framework for efficient AMP generation. Additionally, our prediction module incorporates pre-training and transfer learning strategies for precise AMP identification and screening. We employ a convolutional neural network for multi-attribute prediction and a reinforcement learning-based iterative optimization strategy to produce diverse AMPs. This framework automates molecule generation, screening, attribute prediction and optimization, thereby advancing AMP research. We have also deployed Diff-AMP on a web server, with code, data and server details available in the Data Availability section.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1093/bib/bbae078
- https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdf
- OA Status
- gold
- Cited By
- 58
- References
- 74
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4392571650
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4392571650Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1093/bib/bbae078Digital Object Identifier
- Title
-
Diff-AMP: tailored designed antimicrobial peptide framework with all-in-one generation, identification, prediction and optimizationWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-01-22Full publication date if available
- Authors
-
Rui Wang, Tao Wang, Linlin Zhuo, Jinhang Wei, Xiangzheng Fu, Quan Zou, Xiaojun YaoList of authors in order
- Landing page
-
https://doi.org/10.1093/bib/bbae078Publisher landing page
- PDF URL
-
https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdfDirect OA link when available
- Concepts
-
Computer science, Identification (biology), Artificial intelligence, Reinforcement learning, Machine learning, Antimicrobial peptides, Source code, Interdependence, Deep learning, Code (set theory), Peptide, Chemistry, Set (abstract data type), Biology, Operating system, Programming language, Law, Political science, Botany, BiochemistryTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
58Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 42, 2024: 16Per-year citation counts (last 5 years)
- References (count)
-
74Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4392571650 |
|---|---|
| doi | https://doi.org/10.1093/bib/bbae078 |
| ids.doi | https://doi.org/10.1093/bib/bbae078 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/38446739 |
| ids.openalex | https://openalex.org/W4392571650 |
| fwci | 41.46946595 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000089882 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Antimicrobial Peptides |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D000596 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Amino Acids |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D000900 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Anti-Bacterial Agents |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D004058 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Diffusion |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D007700 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Kinetics |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D000089882 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Antimicrobial Peptides |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D000596 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Amino Acids |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D000900 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Anti-Bacterial Agents |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D004058 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Diffusion |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D007700 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Kinetics |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D000089882 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Antimicrobial Peptides |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000596 |
| mesh[11].is_major_topic | True |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Amino Acids |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D000900 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Anti-Bacterial Agents |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D004058 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Diffusion |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D007700 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Kinetics |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D000089882 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Antimicrobial Peptides |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D000596 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Amino Acids |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D000900 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Anti-Bacterial Agents |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D004058 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Diffusion |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D007700 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Kinetics |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D000089882 |
| mesh[20].is_major_topic | True |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Antimicrobial Peptides |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D000596 |
| mesh[21].is_major_topic | True |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Amino Acids |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D000900 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Anti-Bacterial Agents |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D004058 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Diffusion |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D007700 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Kinetics |
| type | article |
| title | Diff-AMP: tailored designed antimicrobial peptide framework with all-in-one generation, identification, prediction and optimization |
| awards[0].id | https://openalex.org/G552737822 |
| awards[0].funder_id | https://openalex.org/F4320321001 |
| awards[0].display_name | |
| awards[0].funder_award_id | 62372158 |
| awards[0].funder_display_name | National Natural Science Foundation of China |
| awards[1].id | https://openalex.org/G7363650575 |
| awards[1].funder_id | https://openalex.org/F4320321001 |
| awards[1].display_name | |
| awards[1].funder_award_id | 62002111 |
| awards[1].funder_display_name | National Natural Science Foundation of China |
| biblio.issue | 2 |
| biblio.volume | 25 |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11103 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2404 |
| topics[0].subfield.display_name | Microbiology |
| topics[0].display_name | Antimicrobial Peptides and Activities |
| topics[1].id | https://openalex.org/T13326 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9945999979972839 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Biochemical and Structural Characterization |
| topics[2].id | https://openalex.org/T10911 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9869999885559082 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Chemical Synthesis and Analysis |
| funders[0].id | https://openalex.org/F4320321001 |
| funders[0].ror | https://ror.org/01h0zpd94 |
| funders[0].display_name | National Natural Science Foundation of China |
| is_xpac | False |
| apc_list.value | 4011 |
| apc_list.currency | USD |
| apc_list.value_usd | 4011 |
| apc_paid.value | 4011 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 4011 |
| concepts[0].id | https://openalex.org/C41008148 |
| concepts[0].level | 0 |
| concepts[0].score | 0.7835765480995178 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[0].display_name | Computer science |
| concepts[1].id | https://openalex.org/C116834253 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6745709180831909 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2039217 |
| concepts[1].display_name | Identification (biology) |
| concepts[2].id | https://openalex.org/C154945302 |
| concepts[2].level | 1 |
| concepts[2].score | 0.6195429563522339 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[2].display_name | Artificial intelligence |
| concepts[3].id | https://openalex.org/C97541855 |
| concepts[3].level | 2 |
| concepts[3].score | 0.5866641402244568 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q830687 |
| concepts[3].display_name | Reinforcement learning |
| concepts[4].id | https://openalex.org/C119857082 |
| concepts[4].level | 1 |
| concepts[4].score | 0.5829490423202515 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[4].display_name | Machine learning |
| concepts[5].id | https://openalex.org/C540938839 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5714223384857178 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1201508 |
| concepts[5].display_name | Antimicrobial peptides |
| concepts[6].id | https://openalex.org/C43126263 |
| concepts[6].level | 2 |
| concepts[6].score | 0.48767197132110596 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q128751 |
| concepts[6].display_name | Source code |
| concepts[7].id | https://openalex.org/C185874996 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4464673697948456 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q269699 |
| concepts[7].display_name | Interdependence |
| concepts[8].id | https://openalex.org/C108583219 |
| concepts[8].level | 2 |
| concepts[8].score | 0.43783992528915405 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q197536 |
| concepts[8].display_name | Deep learning |
| concepts[9].id | https://openalex.org/C2776760102 |
| concepts[9].level | 3 |
| concepts[9].score | 0.4323325455188751 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q5139990 |
| concepts[9].display_name | Code (set theory) |
| concepts[10].id | https://openalex.org/C2779281246 |
| concepts[10].level | 2 |
| concepts[10].score | 0.15957581996917725 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q172847 |
| concepts[10].display_name | Peptide |
| concepts[11].id | https://openalex.org/C185592680 |
| concepts[11].level | 0 |
| concepts[11].score | 0.10113883018493652 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[11].display_name | Chemistry |
| concepts[12].id | https://openalex.org/C177264268 |
| concepts[12].level | 2 |
| concepts[12].score | 0.09869298338890076 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q1514741 |
| concepts[12].display_name | Set (abstract data type) |
| concepts[13].id | https://openalex.org/C86803240 |
| concepts[13].level | 0 |
| concepts[13].score | 0.08063611388206482 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[13].display_name | Biology |
| concepts[14].id | https://openalex.org/C111919701 |
| concepts[14].level | 1 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q9135 |
| concepts[14].display_name | Operating system |
| concepts[15].id | https://openalex.org/C199360897 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[15].display_name | Programming language |
| concepts[16].id | https://openalex.org/C199539241 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7748 |
| concepts[16].display_name | Law |
| concepts[17].id | https://openalex.org/C17744445 |
| concepts[17].level | 0 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q36442 |
| concepts[17].display_name | Political science |
| concepts[18].id | https://openalex.org/C59822182 |
| concepts[18].level | 1 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q441 |
| concepts[18].display_name | Botany |
| concepts[19].id | https://openalex.org/C55493867 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[19].display_name | Biochemistry |
| keywords[0].id | https://openalex.org/keywords/computer-science |
| keywords[0].score | 0.7835765480995178 |
| keywords[0].display_name | Computer science |
| keywords[1].id | https://openalex.org/keywords/identification |
| keywords[1].score | 0.6745709180831909 |
| keywords[1].display_name | Identification (biology) |
| keywords[2].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[2].score | 0.6195429563522339 |
| keywords[2].display_name | Artificial intelligence |
| keywords[3].id | https://openalex.org/keywords/reinforcement-learning |
| keywords[3].score | 0.5866641402244568 |
| keywords[3].display_name | Reinforcement learning |
| keywords[4].id | https://openalex.org/keywords/machine-learning |
| keywords[4].score | 0.5829490423202515 |
| keywords[4].display_name | Machine learning |
| keywords[5].id | https://openalex.org/keywords/antimicrobial-peptides |
| keywords[5].score | 0.5714223384857178 |
| keywords[5].display_name | Antimicrobial peptides |
| keywords[6].id | https://openalex.org/keywords/source-code |
| keywords[6].score | 0.48767197132110596 |
| keywords[6].display_name | Source code |
| keywords[7].id | https://openalex.org/keywords/interdependence |
| keywords[7].score | 0.4464673697948456 |
| keywords[7].display_name | Interdependence |
| keywords[8].id | https://openalex.org/keywords/deep-learning |
| keywords[8].score | 0.43783992528915405 |
| keywords[8].display_name | Deep learning |
| keywords[9].id | https://openalex.org/keywords/code |
| keywords[9].score | 0.4323325455188751 |
| keywords[9].display_name | Code (set theory) |
| keywords[10].id | https://openalex.org/keywords/peptide |
| keywords[10].score | 0.15957581996917725 |
| keywords[10].display_name | Peptide |
| keywords[11].id | https://openalex.org/keywords/chemistry |
| keywords[11].score | 0.10113883018493652 |
| keywords[11].display_name | Chemistry |
| keywords[12].id | https://openalex.org/keywords/set |
| keywords[12].score | 0.09869298338890076 |
| keywords[12].display_name | Set (abstract data type) |
| keywords[13].id | https://openalex.org/keywords/biology |
| keywords[13].score | 0.08063611388206482 |
| keywords[13].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1093/bib/bbae078 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S91767247 |
| locations[0].source.issn | 1467-5463, 1477-4054 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1467-5463 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Briefings in Bioinformatics |
| locations[0].source.host_organization | https://openalex.org/P4310311648 |
| locations[0].source.host_organization_name | Oxford University Press |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| locations[0].source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Briefings in Bioinformatics |
| locations[0].landing_page_url | https://doi.org/10.1093/bib/bbae078 |
| locations[1].id | pmid:38446739 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Briefings in bioinformatics |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/38446739 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:10939340 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | https://pmc.ncbi.nlm.nih.gov/articles/PMC10939340/pdf/bbae078.pdf |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Brief Bioinform |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/10939340 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5100431117 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Rui Wang |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I146620803, https://openalex.org/I4400573270 |
| authorships[0].affiliations[0].raw_affiliation_string | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[0].institutions[0].id | https://openalex.org/I4400573270 |
| authorships[0].institutions[0].ror | https://ror.org/03dd7qj98 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I4400573270 |
| authorships[0].institutions[0].country_code | |
| authorships[0].institutions[0].display_name | Wenzhou University of Technology |
| authorships[0].institutions[1].id | https://openalex.org/I146620803 |
| authorships[0].institutions[1].ror | https://ror.org/020hxh324 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I146620803 |
| authorships[0].institutions[1].country_code | CN |
| authorships[0].institutions[1].display_name | Wenzhou University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Rui Wang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[1].author.id | https://openalex.org/A5100453693 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3111-2024 |
| authorships[1].author.display_name | Tao Wang |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I146620803, https://openalex.org/I4400573270 |
| authorships[1].affiliations[0].raw_affiliation_string | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[1].institutions[0].id | https://openalex.org/I4400573270 |
| authorships[1].institutions[0].ror | https://ror.org/03dd7qj98 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I4400573270 |
| authorships[1].institutions[0].country_code | |
| authorships[1].institutions[0].display_name | Wenzhou University of Technology |
| authorships[1].institutions[1].id | https://openalex.org/I146620803 |
| authorships[1].institutions[1].ror | https://ror.org/020hxh324 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I146620803 |
| authorships[1].institutions[1].country_code | CN |
| authorships[1].institutions[1].display_name | Wenzhou University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Tao Wang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[2].author.id | https://openalex.org/A5004683765 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-6586-0533 |
| authorships[2].author.display_name | Linlin Zhuo |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I146620803, https://openalex.org/I4400573270 |
| authorships[2].affiliations[0].raw_affiliation_string | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[2].institutions[0].id | https://openalex.org/I4400573270 |
| authorships[2].institutions[0].ror | https://ror.org/03dd7qj98 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I4400573270 |
| authorships[2].institutions[0].country_code | |
| authorships[2].institutions[0].display_name | Wenzhou University of Technology |
| authorships[2].institutions[1].id | https://openalex.org/I146620803 |
| authorships[2].institutions[1].ror | https://ror.org/020hxh324 |
| authorships[2].institutions[1].type | education |
| authorships[2].institutions[1].lineage | https://openalex.org/I146620803 |
| authorships[2].institutions[1].country_code | CN |
| authorships[2].institutions[1].display_name | Wenzhou University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Linlin Zhuo |
| authorships[2].is_corresponding | True |
| authorships[2].raw_affiliation_strings | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[3].author.id | https://openalex.org/A5025402144 |
| authorships[3].author.orcid | https://orcid.org/0009-0003-6957-3472 |
| authorships[3].author.display_name | Jinhang Wei |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I146620803, https://openalex.org/I4400573270 |
| authorships[3].affiliations[0].raw_affiliation_string | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[3].institutions[0].id | https://openalex.org/I4400573270 |
| authorships[3].institutions[0].ror | https://ror.org/03dd7qj98 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I4400573270 |
| authorships[3].institutions[0].country_code | |
| authorships[3].institutions[0].display_name | Wenzhou University of Technology |
| authorships[3].institutions[1].id | https://openalex.org/I146620803 |
| authorships[3].institutions[1].ror | https://ror.org/020hxh324 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I146620803 |
| authorships[3].institutions[1].country_code | CN |
| authorships[3].institutions[1].display_name | Wenzhou University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jinhang Wei |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | School of Data Science and Artificial Intelligence, Wenzhou University of Technology , 325000 Wenzhou , China |
| authorships[4].author.id | https://openalex.org/A5044283271 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-6840-2573 |
| authorships[4].author.display_name | Xiangzheng Fu |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I16609230 |
| authorships[4].affiliations[0].raw_affiliation_string | College of Computer Science and Electronic Engineering, Hunan University , 410012 Changsha , China |
| authorships[4].institutions[0].id | https://openalex.org/I16609230 |
| authorships[4].institutions[0].ror | https://ror.org/05htk5m33 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I16609230 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Hunan University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Xiangzheng Fu |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | College of Computer Science and Electronic Engineering, Hunan University , 410012 Changsha , China |
| authorships[5].author.id | https://openalex.org/A5017426085 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-6406-1142 |
| authorships[5].author.display_name | Quan Zou |
| authorships[5].countries | CN |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I150229711 |
| authorships[5].affiliations[0].raw_affiliation_string | Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China , 611730 Chengdu , China |
| authorships[5].institutions[0].id | https://openalex.org/I150229711 |
| authorships[5].institutions[0].ror | https://ror.org/04qr3zq92 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I150229711 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | University of Electronic Science and Technology of China |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Quan Zou |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China , 611730 Chengdu , China |
| authorships[6].author.id | https://openalex.org/A5101655869 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-9958-8438 |
| authorships[6].author.display_name | Xiaojun Yao |
| authorships[6].countries | MO |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I49835588 |
| authorships[6].affiliations[0].raw_affiliation_string | Faculty of Applied Sciences, Macao Polytechnic University , 999078 Macao , China |
| authorships[6].institutions[0].id | https://openalex.org/I49835588 |
| authorships[6].institutions[0].ror | https://ror.org/02sf5td35 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I49835588 |
| authorships[6].institutions[0].country_code | MO |
| authorships[6].institutions[0].display_name | Macao Polytechnic University |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Xiaojun Yao |
| authorships[6].is_corresponding | True |
| authorships[6].raw_affiliation_strings | Faculty of Applied Sciences, Macao Polytechnic University , 999078 Macao , China |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Diff-AMP: tailored designed antimicrobial peptide framework with all-in-one generation, identification, prediction and optimization |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11103 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2404 |
| primary_topic.subfield.display_name | Microbiology |
| primary_topic.display_name | Antimicrobial Peptides and Activities |
| related_works | https://openalex.org/W2139079562, https://openalex.org/W2749324135, https://openalex.org/W2908433117, https://openalex.org/W2969604939, https://openalex.org/W2378306841, https://openalex.org/W114687057, https://openalex.org/W3122988618, https://openalex.org/W1994631104, https://openalex.org/W4306904969, https://openalex.org/W3146908316 |
| cited_by_count | 58 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 42 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 16 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1093/bib/bbae078 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S91767247 |
| best_oa_location.source.issn | 1467-5463, 1477-4054 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1467-5463 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Briefings in Bioinformatics |
| best_oa_location.source.host_organization | https://openalex.org/P4310311648 |
| best_oa_location.source.host_organization_name | Oxford University Press |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| best_oa_location.source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Briefings in Bioinformatics |
| best_oa_location.landing_page_url | https://doi.org/10.1093/bib/bbae078 |
| primary_location.id | doi:10.1093/bib/bbae078 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S91767247 |
| primary_location.source.issn | 1467-5463, 1477-4054 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1467-5463 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Briefings in Bioinformatics |
| primary_location.source.host_organization | https://openalex.org/P4310311648 |
| primary_location.source.host_organization_name | Oxford University Press |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310311648, https://openalex.org/P4310311647 |
| primary_location.source.host_organization_lineage_names | Oxford University Press, University of Oxford |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://academic.oup.com/bib/article-pdf/25/2/bbae078/56858804/bbae078.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Briefings in Bioinformatics |
| primary_location.landing_page_url | https://doi.org/10.1093/bib/bbae078 |
| publication_date | 2024-01-22 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2092643722, https://openalex.org/W6686723365, https://openalex.org/W1997152791, https://openalex.org/W2097099115, https://openalex.org/W3023413280, https://openalex.org/W2054602599, https://openalex.org/W2058037164, https://openalex.org/W2066275795, https://openalex.org/W2739980134, https://openalex.org/W2037178359, https://openalex.org/W2163251457, https://openalex.org/W2123696501, https://openalex.org/W2084845164, https://openalex.org/W2000788400, https://openalex.org/W3172532482, https://openalex.org/W3177828909, https://openalex.org/W3096831136, https://openalex.org/W6640963894, https://openalex.org/W4324344300, https://openalex.org/W2913423296, https://openalex.org/W4214943162, https://openalex.org/W3145130117, https://openalex.org/W3081615418, https://openalex.org/W3027325047, https://openalex.org/W3045294277, https://openalex.org/W4308052938, https://openalex.org/W3164453494, https://openalex.org/W3025501158, https://openalex.org/W4309578061, https://openalex.org/W2102786297, https://openalex.org/W2059971420, https://openalex.org/W2083501076, https://openalex.org/W2791848964, https://openalex.org/W2194775991, https://openalex.org/W6637373629, https://openalex.org/W3217347090, https://openalex.org/W4308370073, https://openalex.org/W4220952154, https://openalex.org/W2141104259, https://openalex.org/W2074726906, https://openalex.org/W4213077304, https://openalex.org/W3034806393, https://openalex.org/W2342838938, https://openalex.org/W3164941572, https://openalex.org/W2086798171, https://openalex.org/W2140501815, https://openalex.org/W6806209084, https://openalex.org/W4327550249, https://openalex.org/W2189116376, https://openalex.org/W4317942129, https://openalex.org/W6766860405, https://openalex.org/W4322718303, https://openalex.org/W4308834846, https://openalex.org/W4382198814, https://openalex.org/W6756979132, https://openalex.org/W3174864715, https://openalex.org/W6779342232, https://openalex.org/W3117992890, https://openalex.org/W3094708881, https://openalex.org/W2902652978, https://openalex.org/W2141682747, https://openalex.org/W1964937304, https://openalex.org/W2917874934, https://openalex.org/W2984592028, https://openalex.org/W2129115651, https://openalex.org/W3216979156, https://openalex.org/W3188372451, https://openalex.org/W1998064689, https://openalex.org/W3154318479, https://openalex.org/W2511835277, https://openalex.org/W4206816857, https://openalex.org/W3135156311, https://openalex.org/W2965612550, https://openalex.org/W4226148785 |
| referenced_works_count | 74 |
| abstract_inverted_index.a | 84, 135, 143, 173 |
| abstract_inverted_index.We | 100, 133, 167 |
| abstract_inverted_index.in | 183 |
| abstract_inverted_index.it | 51 |
| abstract_inverted_index.of | 18 |
| abstract_inverted_index.on | 172 |
| abstract_inverted_index.to | 23, 28, 70, 149 |
| abstract_inverted_index.we | 82 |
| abstract_inverted_index.AMP | 49, 57, 92, 115, 129, 165 |
| abstract_inverted_index.Due | 27 |
| abstract_inverted_index.The | 15 |
| abstract_inverted_index.and | 11, 33, 78, 97, 105, 123, 131, 142, 161, 179 |
| abstract_inverted_index.are | 36 |
| abstract_inverted_index.for | 40, 113, 127, 139 |
| abstract_inverted_index.has | 21 |
| abstract_inverted_index.led | 22 |
| abstract_inverted_index.low | 30 |
| abstract_inverted_index.our | 118 |
| abstract_inverted_index.the | 60, 109, 184 |
| abstract_inverted_index.web | 174 |
| abstract_inverted_index.AMPs | 35 |
| abstract_inverted_index.Data | 185 |
| abstract_inverted_index.This | 153 |
| abstract_inverted_index.also | 52, 169 |
| abstract_inverted_index.data | 178 |
| abstract_inverted_index.deep | 45, 86 |
| abstract_inverted_index.drug | 31 |
| abstract_inverted_index.fail | 69 |
| abstract_inverted_index.have | 168 |
| abstract_inverted_index.into | 108 |
| abstract_inverted_index.like | 74 |
| abstract_inverted_index.that | 90 |
| abstract_inverted_index.with | 6, 176 |
| abstract_inverted_index.AMPs. | 152 |
| abstract_inverted_index.While | 43 |
| abstract_inverted_index.amino | 64 |
| abstract_inverted_index.among | 63 |
| abstract_inverted_index.code, | 177 |
| abstract_inverted_index.short | 4 |
| abstract_inverted_index.tasks | 73 |
| abstract_inverted_index.their | 29 |
| abstract_inverted_index.acids. | 65 |
| abstract_inverted_index.combat | 12 |
| abstract_inverted_index.employ | 134 |
| abstract_inverted_index.misuse | 17 |
| abstract_inverted_index.models | 68 |
| abstract_inverted_index.module | 120 |
| abstract_inverted_index.neural | 137 |
| abstract_inverted_index.server | 180 |
| abstract_inverted_index.target | 10 |
| abstract_inverted_index.(AMPs), | 3 |
| abstract_inverted_index.certain | 54 |
| abstract_inverted_index.complex | 61 |
| abstract_inverted_index.crucial | 72 |
| abstract_inverted_index.current | 67 |
| abstract_inverted_index.details | 181 |
| abstract_inverted_index.develop | 83 |
| abstract_inverted_index.diverse | 7, 151 |
| abstract_inverted_index.kinetic | 103 |
| abstract_inverted_index.network | 138 |
| abstract_inverted_index.precise | 128 |
| abstract_inverted_index.produce | 150 |
| abstract_inverted_index.server, | 175 |
| abstract_inverted_index.thereby | 163 |
| abstract_inverted_index.various | 13 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Diff-AMP | 171 |
| abstract_inverted_index.Firstly, | 56 |
| abstract_inverted_index.chemical | 19 |
| abstract_inverted_index.deployed | 170 |
| abstract_inverted_index.enhances | 48 |
| abstract_inverted_index.existing | 44 |
| abstract_inverted_index.learning | 46, 87, 111, 125 |
| abstract_inverted_index.molecule | 156 |
| abstract_inverted_index.peptides | 2, 5 |
| abstract_inverted_index.presents | 53 |
| abstract_inverted_index.section. | 187 |
| abstract_inverted_index.strategy | 148 |
| abstract_inverted_index.transfer | 124 |
| abstract_inverted_index.Diff-AMP, | 89 |
| abstract_inverted_index.Secondly, | 66 |
| abstract_inverted_index.advancing | 164 |
| abstract_inverted_index.attention | 106 |
| abstract_inverted_index.attribute | 76, 95, 159 |
| abstract_inverted_index.automates | 91, 155 |
| abstract_inverted_index.available | 182 |
| abstract_inverted_index.diffusion | 104 |
| abstract_inverted_index.efficient | 114 |
| abstract_inverted_index.framework | 112, 154 |
| abstract_inverted_index.integrate | 71, 102 |
| abstract_inverted_index.iterative | 79, 98, 146 |
| abstract_inverted_index.microbial | 25 |
| abstract_inverted_index.overlooks | 59 |
| abstract_inverted_index.promising | 38 |
| abstract_inverted_index.research. | 166 |
| abstract_inverted_index.toxicity, | 34 |
| abstract_inverted_index.considered | 37 |
| abstract_inverted_index.framework, | 88 |
| abstract_inverted_index.functions, | 8 |
| abstract_inverted_index.generation | 58 |
| abstract_inverted_index.increasing | 24 |
| abstract_inverted_index.integrated | 85 |
| abstract_inverted_index.mechanisms | 107 |
| abstract_inverted_index.organisms. | 14 |
| abstract_inverted_index.prediction | 77, 96, 119, 141, 160 |
| abstract_inverted_index.resistance | 32 |
| abstract_inverted_index.screening, | 75, 158 |
| abstract_inverted_index.screening. | 132 |
| abstract_inverted_index.strategies | 126 |
| abstract_inverted_index.technology | 47 |
| abstract_inverted_index.widespread | 16 |
| abstract_inverted_index.antibiotics | 20 |
| abstract_inverted_index.challenges. | 55 |
| abstract_inverted_index.effectively | 9 |
| abstract_inverted_index.generation, | 50, 93, 157 |
| abstract_inverted_index.generation. | 116 |
| abstract_inverted_index.resistance. | 26 |
| abstract_inverted_index.substitutes | 39 |
| abstract_inverted_index.traditional | 41 |
| abstract_inverted_index.Availability | 186 |
| abstract_inverted_index.antibiotics. | 42 |
| abstract_inverted_index.incorporates | 121 |
| abstract_inverted_index.innovatively | 101 |
| abstract_inverted_index.optimization | 147 |
| abstract_inverted_index.pre-training | 122 |
| abstract_inverted_index.Additionally, | 117 |
| abstract_inverted_index.Antimicrobial | 1 |
| abstract_inverted_index.Consequently, | 81 |
| abstract_inverted_index.convolutional | 136 |
| abstract_inverted_index.optimization, | 162 |
| abstract_inverted_index.optimization. | 80, 99 |
| abstract_inverted_index.reinforcement | 110, 144 |
| abstract_inverted_index.identification | 130 |
| abstract_inverted_index.learning-based | 145 |
| abstract_inverted_index.identification, | 94 |
| abstract_inverted_index.multi-attribute | 140 |
| abstract_inverted_index.interdependencies | 62 |
| cited_by_percentile_year.max | 100 |
| cited_by_percentile_year.min | 99 |
| corresponding_author_ids | https://openalex.org/A5101655869, https://openalex.org/A5044283271, https://openalex.org/A5004683765, https://openalex.org/A5017426085 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I146620803, https://openalex.org/I150229711, https://openalex.org/I16609230, https://openalex.org/I4400573270, https://openalex.org/I49835588 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.5799999833106995 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.99837009 |
| citation_normalized_percentile.is_in_top_1_percent | True |
| citation_normalized_percentile.is_in_top_10_percent | True |