Discovery and functional annotation of quantitative trait loci affecting resistance to sea lice in Atlantic salmon Article Swipe
 YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1101/455626
YOU?
·
· 2018
· Open Access
·
· DOI: https://doi.org/10.1101/455626
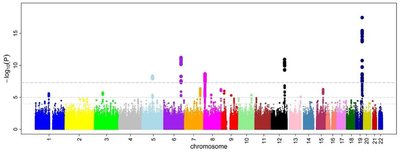
Sea lice ( Caligus rogercresseyi ) are ectoparasitic copepods which have a large negative economic and welfare impact in Atlantic salmon ( Salmo salar ) aquaculture, particularly in Chile. A multi-faceted prevention and control strategy is required to tackle lice, and selective breeding contributes via cumulative improvement of host resistance to the parasite. While host resistance has been shown to be heritable, little is yet known about the individual loci that contribute to this resistance, the potential underlying genes, and their mechanisms of action. In this study we took a multifaceted approach to identify and characterise quantitative trait loci (QTL) affecting hose resistance in a population of 2,688 Caligus-challenged Atlantic salmon post-smolts from a commercial breeding programme. We used low and medium density genotyping to collect genome-wide SNP marker data for all animals. Moderate heritablility estimates of 0.28 and 0.24 were obtained for lice density (as a measure of host resistance) and growth during infestation respectively. Three QTL explaining between 7 and 13 % of the genetic variation in resistance to sea lice (as represented by the traits of lice density) were detected on chromosomes 3, 18 and 21. Characterisation of these QTL regions was undertaken using RNA sequencing and pooled whole genome sequencing data. This resulted in the identification of a shortlist of potential underlying causative genes, and candidate functional mutations for further study. For example, candidates within the chromosome 3 QTL include a putative premature stop mutation in TOB1 (an anti-proliferative transcription factor involved in T cell regulation) and an uncharacterized protein which showed significant differential allelic expression (implying the existence of a cis-acting regulatory mutation). While host resistance to sea lice is polygenic in nature, the results of this study highlight significant QTL regions together explaining a moderate proportion of the heritability of the trait. Future investigation of these QTL may enable improved knowledge of the functional mechanisms of host resistance to sea lice, and incorporation of functional variants to improve genomic selection accuracy.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/455626
- https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdf
- OA Status
- green
- Cited By
- 4
- References
- 58
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2952197921
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2952197921Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/455626Digital Object Identifier
- Title
-
Discovery and functional annotation of quantitative trait loci affecting resistance to sea lice in Atlantic salmonWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2018Year of publication
- Publication date
-
2018-10-29Full publication date if available
- Authors
-
Diego Robledo, Alejandro P. Gutiérrez, Agustín Barría, Jean P. Lhorente, Ross D. Houston, José M. YáñezList of authors in order
- Landing page
-
https://doi.org/10.1101/455626Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdfDirect OA link when available
- Concepts
-
Biology, Quantitative trait locus, Salmo, Genetics, Lepeophtheirus, Genotyping, Candidate gene, Gene, Single-nucleotide polymorphism, Genome-wide association study, Genotype, Fishery, FishTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
4Total citation count in OpenAlex
- Citations by year (recent)
-
2023: 1, 2022: 1, 2021: 1, 2019: 1Per-year citation counts (last 5 years)
- References (count)
-
58Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2952197921 |
|---|---|
| doi | https://doi.org/10.1101/455626 |
| ids.doi | https://doi.org/10.1101/455626 |
| ids.mag | 2952197921 |
| ids.openalex | https://openalex.org/W2952197921 |
| fwci | 0.35314015 |
| type | preprint |
| title | Discovery and functional annotation of quantitative trait loci affecting resistance to sea lice in Atlantic salmon |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10012 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9991000294685364 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic diversity and population structure |
| topics[1].id | https://openalex.org/T10594 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9984999895095825 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Genetic and phenotypic traits in livestock |
| topics[2].id | https://openalex.org/T10506 |
| topics[2].field.id | https://openalex.org/fields/24 |
| topics[2].field.display_name | Immunology and Microbiology |
| topics[2].score | 0.9970999956130981 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2403 |
| topics[2].subfield.display_name | Immunology |
| topics[2].display_name | Aquaculture disease management and microbiota |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C86803240 |
| concepts[0].level | 0 |
| concepts[0].score | 0.9137846231460571 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[0].display_name | Biology |
| concepts[1].id | https://openalex.org/C81941488 |
| concepts[1].level | 3 |
| concepts[1].score | 0.9080429077148438 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q853421 |
| concepts[1].display_name | Quantitative trait locus |
| concepts[2].id | https://openalex.org/C2777940460 |
| concepts[2].level | 3 |
| concepts[2].score | 0.7588930130004883 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q310436 |
| concepts[2].display_name | Salmo |
| concepts[3].id | https://openalex.org/C54355233 |
| concepts[3].level | 1 |
| concepts[3].score | 0.6727806925773621 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[3].display_name | Genetics |
| concepts[4].id | https://openalex.org/C2780253540 |
| concepts[4].level | 4 |
| concepts[4].score | 0.598613977432251 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q4286834 |
| concepts[4].display_name | Lepeophtheirus |
| concepts[5].id | https://openalex.org/C31467283 |
| concepts[5].level | 4 |
| concepts[5].score | 0.5380546450614929 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q912147 |
| concepts[5].display_name | Genotyping |
| concepts[6].id | https://openalex.org/C69991583 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4600209593772888 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1423575 |
| concepts[6].display_name | Candidate gene |
| concepts[7].id | https://openalex.org/C104317684 |
| concepts[7].level | 2 |
| concepts[7].score | 0.45684799551963806 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[7].display_name | Gene |
| concepts[8].id | https://openalex.org/C153209595 |
| concepts[8].level | 4 |
| concepts[8].score | 0.4347427487373352 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[8].display_name | Single-nucleotide polymorphism |
| concepts[9].id | https://openalex.org/C106208931 |
| concepts[9].level | 5 |
| concepts[9].score | 0.4242209792137146 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[9].display_name | Genome-wide association study |
| concepts[10].id | https://openalex.org/C135763542 |
| concepts[10].level | 3 |
| concepts[10].score | 0.2811738848686218 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[10].display_name | Genotype |
| concepts[11].id | https://openalex.org/C505870484 |
| concepts[11].level | 1 |
| concepts[11].score | 0.22514033317565918 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q180538 |
| concepts[11].display_name | Fishery |
| concepts[12].id | https://openalex.org/C2909208804 |
| concepts[12].level | 2 |
| concepts[12].score | 0.12444308400154114 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q127282 |
| concepts[12].display_name | Fish |
| keywords[0].id | https://openalex.org/keywords/biology |
| keywords[0].score | 0.9137846231460571 |
| keywords[0].display_name | Biology |
| keywords[1].id | https://openalex.org/keywords/quantitative-trait-locus |
| keywords[1].score | 0.9080429077148438 |
| keywords[1].display_name | Quantitative trait locus |
| keywords[2].id | https://openalex.org/keywords/salmo |
| keywords[2].score | 0.7588930130004883 |
| keywords[2].display_name | Salmo |
| keywords[3].id | https://openalex.org/keywords/genetics |
| keywords[3].score | 0.6727806925773621 |
| keywords[3].display_name | Genetics |
| keywords[4].id | https://openalex.org/keywords/lepeophtheirus |
| keywords[4].score | 0.598613977432251 |
| keywords[4].display_name | Lepeophtheirus |
| keywords[5].id | https://openalex.org/keywords/genotyping |
| keywords[5].score | 0.5380546450614929 |
| keywords[5].display_name | Genotyping |
| keywords[6].id | https://openalex.org/keywords/candidate-gene |
| keywords[6].score | 0.4600209593772888 |
| keywords[6].display_name | Candidate gene |
| keywords[7].id | https://openalex.org/keywords/gene |
| keywords[7].score | 0.45684799551963806 |
| keywords[7].display_name | Gene |
| keywords[8].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[8].score | 0.4347427487373352 |
| keywords[8].display_name | Single-nucleotide polymorphism |
| keywords[9].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[9].score | 0.4242209792137146 |
| keywords[9].display_name | Genome-wide association study |
| keywords[10].id | https://openalex.org/keywords/genotype |
| keywords[10].score | 0.2811738848686218 |
| keywords[10].display_name | Genotype |
| keywords[11].id | https://openalex.org/keywords/fishery |
| keywords[11].score | 0.22514033317565918 |
| keywords[11].display_name | Fishery |
| keywords[12].id | https://openalex.org/keywords/fish-actinopterygii |
| keywords[12].score | 0.12444308400154114 |
| keywords[12].display_name | Fish |
| language | en |
| locations[0].id | doi:10.1101/455626 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/455626 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5071973746 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-9616-5912 |
| authorships[0].author.display_name | Diego Robledo |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I133960621, https://openalex.org/I98677209 |
| authorships[0].affiliations[0].raw_affiliation_string | The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK |
| authorships[0].institutions[0].id | https://openalex.org/I133960621 |
| authorships[0].institutions[0].ror | https://ror.org/01920rj20 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I133960621, https://openalex.org/I2799693246, https://openalex.org/I4210087105, https://openalex.org/I98677209 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | Roslin Institute |
| authorships[0].institutions[1].id | https://openalex.org/I98677209 |
| authorships[0].institutions[1].ror | https://ror.org/01nrxwf90 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I98677209 |
| authorships[0].institutions[1].country_code | GB |
| authorships[0].institutions[1].display_name | University of Edinburgh |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Diego Robledo |
| authorships[0].is_corresponding | True |
| authorships[0].raw_affiliation_strings | The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK |
| authorships[1].author.id | https://openalex.org/A5007562201 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3952-2896 |
| authorships[1].author.display_name | Alejandro P. Gutiérrez |
| authorships[1].countries | GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I133960621, https://openalex.org/I98677209 |
| authorships[1].affiliations[0].raw_affiliation_string | The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK |
| authorships[1].institutions[0].id | https://openalex.org/I133960621 |
| authorships[1].institutions[0].ror | https://ror.org/01920rj20 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I133960621, https://openalex.org/I2799693246, https://openalex.org/I4210087105, https://openalex.org/I98677209 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | Roslin Institute |
| authorships[1].institutions[1].id | https://openalex.org/I98677209 |
| authorships[1].institutions[1].ror | https://ror.org/01nrxwf90 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I98677209 |
| authorships[1].institutions[1].country_code | GB |
| authorships[1].institutions[1].display_name | University of Edinburgh |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Alejandro P. Gutiérrez |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK |
| authorships[2].author.id | https://openalex.org/A5079000726 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-2813-4559 |
| authorships[2].author.display_name | Agustín Barría |
| authorships[2].countries | CL |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I69737025 |
| authorships[2].affiliations[0].raw_affiliation_string | Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santiago, Chile |
| authorships[2].institutions[0].id | https://openalex.org/I69737025 |
| authorships[2].institutions[0].ror | https://ror.org/047gc3g35 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I69737025 |
| authorships[2].institutions[0].country_code | CL |
| authorships[2].institutions[0].display_name | University of Chile |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Agustín Barría |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santiago, Chile |
| authorships[3].author.id | https://openalex.org/A5088148607 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-9157-4231 |
| authorships[3].author.display_name | Jean P. Lhorente |
| authorships[3].affiliations[0].raw_affiliation_string | Benchmark Genetics Chile, Puerto Montt, Chile |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jean P. Lhorente |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Benchmark Genetics Chile, Puerto Montt, Chile |
| authorships[4].author.id | https://openalex.org/A5005051455 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-1805-0762 |
| authorships[4].author.display_name | Ross D. Houston |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I133960621, https://openalex.org/I98677209 |
| authorships[4].affiliations[0].raw_affiliation_string | The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK |
| authorships[4].institutions[0].id | https://openalex.org/I133960621 |
| authorships[4].institutions[0].ror | https://ror.org/01920rj20 |
| authorships[4].institutions[0].type | facility |
| authorships[4].institutions[0].lineage | https://openalex.org/I133960621, https://openalex.org/I2799693246, https://openalex.org/I4210087105, https://openalex.org/I98677209 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | Roslin Institute |
| authorships[4].institutions[1].id | https://openalex.org/I98677209 |
| authorships[4].institutions[1].ror | https://ror.org/01nrxwf90 |
| authorships[4].institutions[1].type | education |
| authorships[4].institutions[1].lineage | https://openalex.org/I98677209 |
| authorships[4].institutions[1].country_code | GB |
| authorships[4].institutions[1].display_name | University of Edinburgh |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Ross D. Houston |
| authorships[4].is_corresponding | True |
| authorships[4].raw_affiliation_strings | The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK |
| authorships[5].author.id | https://openalex.org/A5048349962 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-6612-4087 |
| authorships[5].author.display_name | José M. Yáñez |
| authorships[5].countries | CL |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I69737025 |
| authorships[5].affiliations[0].raw_affiliation_string | Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santiago, Chile |
| authorships[5].affiliations[1].raw_affiliation_string | Benchmark Genetics Chile, Puerto Montt, Chile |
| authorships[5].affiliations[2].institution_ids | https://openalex.org/I4210156632 |
| authorships[5].affiliations[2].raw_affiliation_string | Núcleo Milenio INVASAL, Concepción, Chile |
| authorships[5].institutions[0].id | https://openalex.org/I4210156632 |
| authorships[5].institutions[0].ror | https://ror.org/054rvnp37 |
| authorships[5].institutions[0].type | other |
| authorships[5].institutions[0].lineage | https://openalex.org/I4210156632 |
| authorships[5].institutions[0].country_code | CL |
| authorships[5].institutions[0].display_name | Millenium Nucleus for Planet Formation |
| authorships[5].institutions[1].id | https://openalex.org/I69737025 |
| authorships[5].institutions[1].ror | https://ror.org/047gc3g35 |
| authorships[5].institutions[1].type | education |
| authorships[5].institutions[1].lineage | https://openalex.org/I69737025 |
| authorships[5].institutions[1].country_code | CL |
| authorships[5].institutions[1].display_name | University of Chile |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | José M. Yáñez |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Benchmark Genetics Chile, Puerto Montt, Chile, Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, Santiago, Chile, Núcleo Milenio INVASAL, Concepción, Chile |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Discovery and functional annotation of quantitative trait loci affecting resistance to sea lice in Atlantic salmon |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10012 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9991000294685364 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic diversity and population structure |
| related_works | https://openalex.org/W2953176467, https://openalex.org/W2025983546, https://openalex.org/W2322357082, https://openalex.org/W3123885297, https://openalex.org/W2794816152, https://openalex.org/W2158988556, https://openalex.org/W2042936342, https://openalex.org/W1986742794, https://openalex.org/W2092286609, https://openalex.org/W4253277708 |
| cited_by_count | 4 |
| counts_by_year[0].year | 2023 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2022 |
| counts_by_year[1].cited_by_count | 1 |
| counts_by_year[2].year | 2021 |
| counts_by_year[2].cited_by_count | 1 |
| counts_by_year[3].year | 2019 |
| counts_by_year[3].cited_by_count | 1 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/455626 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/455626 |
| primary_location.id | doi:10.1101/455626 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2018/10/29/455626.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/455626 |
| publication_date | 2018-10-29 |
| publication_year | 2018 |
| referenced_works | https://openalex.org/W2010873394, https://openalex.org/W1993616000, https://openalex.org/W2134432161, https://openalex.org/W2161245845, https://openalex.org/W1997410198, https://openalex.org/W2131271579, https://openalex.org/W1998181451, https://openalex.org/W2076620170, https://openalex.org/W2149378411, https://openalex.org/W2148396262, https://openalex.org/W2341151106, https://openalex.org/W2131693276, https://openalex.org/W2116056560, https://openalex.org/W2169456326, https://openalex.org/W2139911862, https://openalex.org/W2095818343, https://openalex.org/W2811205482, https://openalex.org/W1491365253, https://openalex.org/W2156764697, https://openalex.org/W2190107622, https://openalex.org/W2081748519, https://openalex.org/W2761275051, https://openalex.org/W2059827462, https://openalex.org/W2740620321, https://openalex.org/W2760980242, https://openalex.org/W1985758132, https://openalex.org/W2005501593, https://openalex.org/W2161950199, https://openalex.org/W2033184271, https://openalex.org/W2789333397, https://openalex.org/W2106265043, https://openalex.org/W2018633460, https://openalex.org/W2108234281, https://openalex.org/W2337283254, https://openalex.org/W2575872105, https://openalex.org/W2271739161, https://openalex.org/W2224056471, https://openalex.org/W2144612407, https://openalex.org/W2075009410, https://openalex.org/W1519622305, https://openalex.org/W2047875731, https://openalex.org/W2053588621, https://openalex.org/W2212983258, https://openalex.org/W1972826622, https://openalex.org/W2791415333, https://openalex.org/W2951568231, https://openalex.org/W2110682604, https://openalex.org/W2471746432, https://openalex.org/W2060933776, https://openalex.org/W2154412838, https://openalex.org/W2590121248, https://openalex.org/W2049569650, https://openalex.org/W2271012763, https://openalex.org/W2010828744, https://openalex.org/W1928998639, https://openalex.org/W2095680943, https://openalex.org/W2199798003, https://openalex.org/W2585769723 |
| referenced_works_count | 58 |
| abstract_inverted_index.% | 164 |
| abstract_inverted_index.( | 3, 22 |
| abstract_inverted_index.) | 6, 25 |
| abstract_inverted_index.3 | 232 |
| abstract_inverted_index.7 | 161 |
| abstract_inverted_index.A | 30 |
| abstract_inverted_index.T | 248 |
| abstract_inverted_index.a | 12, 90, 105, 114, 147, 212, 235, 265, 290 |
| abstract_inverted_index.13 | 163 |
| abstract_inverted_index.18 | 187 |
| abstract_inverted_index.3, | 186 |
| abstract_inverted_index.In | 85 |
| abstract_inverted_index.We | 118 |
| abstract_inverted_index.an | 252 |
| abstract_inverted_index.be | 61 |
| abstract_inverted_index.by | 176 |
| abstract_inverted_index.in | 19, 28, 104, 169, 208, 240, 247, 277 |
| abstract_inverted_index.is | 36, 64, 275 |
| abstract_inverted_index.of | 48, 83, 107, 137, 149, 165, 179, 191, 211, 214, 264, 281, 293, 296, 301, 308, 312, 320 |
| abstract_inverted_index.on | 184 |
| abstract_inverted_index.to | 38, 51, 60, 73, 93, 125, 171, 272, 315, 323 |
| abstract_inverted_index.we | 88 |
| abstract_inverted_index.(an | 242 |
| abstract_inverted_index.(as | 146, 174 |
| abstract_inverted_index.21. | 189 |
| abstract_inverted_index.For | 226 |
| abstract_inverted_index.QTL | 158, 193, 233, 286, 303 |
| abstract_inverted_index.RNA | 198 |
| abstract_inverted_index.SNP | 128 |
| abstract_inverted_index.Sea | 1 |
| abstract_inverted_index.all | 132 |
| abstract_inverted_index.and | 16, 33, 41, 80, 95, 121, 139, 152, 162, 188, 200, 219, 251, 318 |
| abstract_inverted_index.are | 7 |
| abstract_inverted_index.for | 131, 143, 223 |
| abstract_inverted_index.has | 57 |
| abstract_inverted_index.low | 120 |
| abstract_inverted_index.may | 304 |
| abstract_inverted_index.sea | 172, 273, 316 |
| abstract_inverted_index.the | 52, 68, 76, 166, 177, 209, 230, 262, 279, 294, 297, 309 |
| abstract_inverted_index.via | 45 |
| abstract_inverted_index.was | 195 |
| abstract_inverted_index.yet | 65 |
| abstract_inverted_index.0.24 | 140 |
| abstract_inverted_index.0.28 | 138 |
| abstract_inverted_index.TOB1 | 241 |
| abstract_inverted_index.This | 206 |
| abstract_inverted_index.been | 58 |
| abstract_inverted_index.cell | 249 |
| abstract_inverted_index.data | 130 |
| abstract_inverted_index.from | 113 |
| abstract_inverted_index.have | 11 |
| abstract_inverted_index.hose | 102 |
| abstract_inverted_index.host | 49, 55, 150, 270, 313 |
| abstract_inverted_index.lice | 2, 144, 173, 180, 274 |
| abstract_inverted_index.loci | 70, 99 |
| abstract_inverted_index.stop | 238 |
| abstract_inverted_index.that | 71 |
| abstract_inverted_index.this | 74, 86, 282 |
| abstract_inverted_index.took | 89 |
| abstract_inverted_index.used | 119 |
| abstract_inverted_index.were | 141, 182 |
| abstract_inverted_index.(QTL) | 100 |
| abstract_inverted_index.2,688 | 108 |
| abstract_inverted_index.Salmo | 23 |
| abstract_inverted_index.Three | 157 |
| abstract_inverted_index.While | 54, 269 |
| abstract_inverted_index.about | 67 |
| abstract_inverted_index.data. | 205 |
| abstract_inverted_index.known | 66 |
| abstract_inverted_index.large | 13 |
| abstract_inverted_index.lice, | 40, 317 |
| abstract_inverted_index.salar | 24 |
| abstract_inverted_index.shown | 59 |
| abstract_inverted_index.study | 87, 283 |
| abstract_inverted_index.their | 81 |
| abstract_inverted_index.these | 192, 302 |
| abstract_inverted_index.trait | 98 |
| abstract_inverted_index.using | 197 |
| abstract_inverted_index.which | 10, 255 |
| abstract_inverted_index.whole | 202 |
| abstract_inverted_index.Chile. | 29 |
| abstract_inverted_index.Future | 299 |
| abstract_inverted_index.during | 154 |
| abstract_inverted_index.enable | 305 |
| abstract_inverted_index.factor | 245 |
| abstract_inverted_index.genes, | 79, 218 |
| abstract_inverted_index.genome | 203 |
| abstract_inverted_index.growth | 153 |
| abstract_inverted_index.impact | 18 |
| abstract_inverted_index.little | 63 |
| abstract_inverted_index.marker | 129 |
| abstract_inverted_index.medium | 122 |
| abstract_inverted_index.pooled | 201 |
| abstract_inverted_index.salmon | 21, 111 |
| abstract_inverted_index.showed | 256 |
| abstract_inverted_index.study. | 225 |
| abstract_inverted_index.tackle | 39 |
| abstract_inverted_index.trait. | 298 |
| abstract_inverted_index.traits | 178 |
| abstract_inverted_index.within | 229 |
| abstract_inverted_index.Caligus | 4 |
| abstract_inverted_index.action. | 84 |
| abstract_inverted_index.allelic | 259 |
| abstract_inverted_index.between | 160 |
| abstract_inverted_index.collect | 126 |
| abstract_inverted_index.control | 34 |
| abstract_inverted_index.density | 123, 145 |
| abstract_inverted_index.further | 224 |
| abstract_inverted_index.genetic | 167 |
| abstract_inverted_index.genomic | 325 |
| abstract_inverted_index.improve | 324 |
| abstract_inverted_index.include | 234 |
| abstract_inverted_index.measure | 148 |
| abstract_inverted_index.nature, | 278 |
| abstract_inverted_index.protein | 254 |
| abstract_inverted_index.regions | 194, 287 |
| abstract_inverted_index.results | 280 |
| abstract_inverted_index.welfare | 17 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.Atlantic | 20, 110 |
| abstract_inverted_index.Moderate | 134 |
| abstract_inverted_index.animals. | 133 |
| abstract_inverted_index.approach | 92 |
| abstract_inverted_index.breeding | 43, 116 |
| abstract_inverted_index.copepods | 9 |
| abstract_inverted_index.density) | 181 |
| abstract_inverted_index.detected | 183 |
| abstract_inverted_index.economic | 15 |
| abstract_inverted_index.example, | 227 |
| abstract_inverted_index.identify | 94 |
| abstract_inverted_index.improved | 306 |
| abstract_inverted_index.involved | 246 |
| abstract_inverted_index.moderate | 291 |
| abstract_inverted_index.mutation | 239 |
| abstract_inverted_index.negative | 14 |
| abstract_inverted_index.obtained | 142 |
| abstract_inverted_index.putative | 236 |
| abstract_inverted_index.required | 37 |
| abstract_inverted_index.resulted | 207 |
| abstract_inverted_index.strategy | 35 |
| abstract_inverted_index.together | 288 |
| abstract_inverted_index.variants | 322 |
| abstract_inverted_index.(implying | 261 |
| abstract_inverted_index.accuracy. | 327 |
| abstract_inverted_index.affecting | 101 |
| abstract_inverted_index.candidate | 220 |
| abstract_inverted_index.causative | 217 |
| abstract_inverted_index.estimates | 136 |
| abstract_inverted_index.existence | 263 |
| abstract_inverted_index.highlight | 284 |
| abstract_inverted_index.knowledge | 307 |
| abstract_inverted_index.mutations | 222 |
| abstract_inverted_index.parasite. | 53 |
| abstract_inverted_index.polygenic | 276 |
| abstract_inverted_index.potential | 77, 215 |
| abstract_inverted_index.premature | 237 |
| abstract_inverted_index.selection | 326 |
| abstract_inverted_index.selective | 42 |
| abstract_inverted_index.shortlist | 213 |
| abstract_inverted_index.variation | 168 |
| abstract_inverted_index.candidates | 228 |
| abstract_inverted_index.chromosome | 231 |
| abstract_inverted_index.cis-acting | 266 |
| abstract_inverted_index.commercial | 115 |
| abstract_inverted_index.contribute | 72 |
| abstract_inverted_index.cumulative | 46 |
| abstract_inverted_index.explaining | 159, 289 |
| abstract_inverted_index.expression | 260 |
| abstract_inverted_index.functional | 221, 310, 321 |
| abstract_inverted_index.genotyping | 124 |
| abstract_inverted_index.heritable, | 62 |
| abstract_inverted_index.individual | 69 |
| abstract_inverted_index.mechanisms | 82, 311 |
| abstract_inverted_index.mutation). | 268 |
| abstract_inverted_index.population | 106 |
| abstract_inverted_index.prevention | 32 |
| abstract_inverted_index.programme. | 117 |
| abstract_inverted_index.proportion | 292 |
| abstract_inverted_index.regulatory | 267 |
| abstract_inverted_index.resistance | 50, 56, 103, 170, 271, 314 |
| abstract_inverted_index.sequencing | 199, 204 |
| abstract_inverted_index.underlying | 78, 216 |
| abstract_inverted_index.undertaken | 196 |
| abstract_inverted_index.chromosomes | 185 |
| abstract_inverted_index.contributes | 44 |
| abstract_inverted_index.genome-wide | 127 |
| abstract_inverted_index.improvement | 47 |
| abstract_inverted_index.infestation | 155 |
| abstract_inverted_index.post-smolts | 112 |
| abstract_inverted_index.regulation) | 250 |
| abstract_inverted_index.represented | 175 |
| abstract_inverted_index.resistance) | 151 |
| abstract_inverted_index.resistance, | 75 |
| abstract_inverted_index.significant | 257, 285 |
| abstract_inverted_index.aquaculture, | 26 |
| abstract_inverted_index.characterise | 96 |
| abstract_inverted_index.differential | 258 |
| abstract_inverted_index.heritability | 295 |
| abstract_inverted_index.multifaceted | 91 |
| abstract_inverted_index.particularly | 27 |
| abstract_inverted_index.quantitative | 97 |
| abstract_inverted_index.ectoparasitic | 8 |
| abstract_inverted_index.heritablility | 135 |
| abstract_inverted_index.incorporation | 319 |
| abstract_inverted_index.investigation | 300 |
| abstract_inverted_index.multi-faceted | 31 |
| abstract_inverted_index.respectively. | 156 |
| abstract_inverted_index.rogercresseyi | 5 |
| abstract_inverted_index.transcription | 244 |
| abstract_inverted_index.identification | 210 |
| abstract_inverted_index.uncharacterized | 253 |
| abstract_inverted_index.Characterisation | 190 |
| abstract_inverted_index.Caligus-challenged | 109 |
| abstract_inverted_index.anti-proliferative | 243 |
| cited_by_percentile_year.max | 94 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5048349962, https://openalex.org/A5071973746, https://openalex.org/A5005051455 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 6 |
| corresponding_institution_ids | https://openalex.org/I133960621, https://openalex.org/I4210156632, https://openalex.org/I69737025, https://openalex.org/I98677209 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/14 |
| sustainable_development_goals[0].score | 0.8799999952316284 |
| sustainable_development_goals[0].display_name | Life below water |
| citation_normalized_percentile.value | 0.70116549 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |