Dissecting signalling hierarchies in the patterning of the mouse primitive streak using micro-patterned EpiLC colonies Article Swipe
 YOU?
·
· 2020
· Open Access
·
· DOI: https://doi.org/10.1101/2020.11.30.404418
YOU?
·
· 2020
· Open Access
·
· DOI: https://doi.org/10.1101/2020.11.30.404418
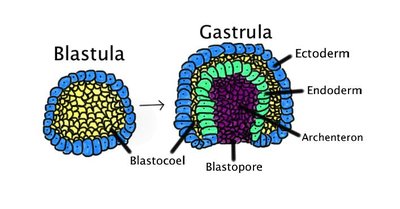
Molecular embryology studies have established that the patterning of the gastrula-stage mouse embryo is dependent on a regulatory network where the WNT, BMP and NODAL signalling pathways cooperate. Still, important aspects of their respective contributions to this process remain unclear. Here, studying their impact on the spatial organization and the developmental trajectories of micro-patterned Epiblast Like Cells (EpiLC) colonies, we show that when BMP is present, it dominates NODAL and WNT and imposes a posterior character to the colonies differentiation. However, the use of two Nodal mutant cell lines allowed us to show that prior to BMP action, NODAL is required to establish the mesendodermal lineage. The fact that mutant phenotypes were more severe in vitro than in vivo suggests that embryonic phenotypes are partially rescued by ligands of extra-embryonic or maternal origin. Our work demonstrates the complementarity of micro-patterned EpiLC colonies to embryological approaches.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2020.11.30.404418
- https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdf
- OA Status
- green
- Cited By
- 1
- References
- 60
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3107164815
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3107164815Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2020.11.30.404418Digital Object Identifier
- Title
-
Dissecting signalling hierarchies in the patterning of the mouse primitive streak using micro-patterned EpiLC coloniesWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2020Year of publication
- Publication date
-
2020-11-30Full publication date if available
- Authors
-
Jean-Louis Plouhinec, Mathieu Vieira, Gaël Simon, Jérôme Collignon, Benoît SorreList of authors in order
- Landing page
-
https://doi.org/10.1101/2020.11.30.404418Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdfDirect OA link when available
- Concepts
-
Gastrulation, Epiblast, Biology, NODAL, Nodal signaling, Embryonic stem cell, Cell biology, Wnt signaling pathway, Primitive streak, Phenotype, Lineage (genetic), Embryo, Endoderm, Genetics, Embryogenesis, Signal transduction, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2022: 1Per-year citation counts (last 5 years)
- References (count)
-
60Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3107164815 |
|---|---|
| doi | https://doi.org/10.1101/2020.11.30.404418 |
| ids.doi | https://doi.org/10.1101/2020.11.30.404418 |
| ids.mag | 3107164815 |
| ids.openalex | https://openalex.org/W3107164815 |
| fwci | 0.09752697 |
| type | preprint |
| title | Dissecting signalling hierarchies in the patterning of the mouse primitive streak using micro-patterned EpiLC colonies |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10505 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Pluripotent Stem Cells Research |
| topics[1].id | https://openalex.org/T11190 |
| topics[1].field.id | https://openalex.org/fields/22 |
| topics[1].field.display_name | Engineering |
| topics[1].score | 0.9980999827384949 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2204 |
| topics[1].subfield.display_name | Biomedical Engineering |
| topics[1].display_name | 3D Printing in Biomedical Research |
| topics[2].id | https://openalex.org/T12264 |
| topics[2].field.id | https://openalex.org/fields/27 |
| topics[2].field.display_name | Medicine |
| topics[2].score | 0.9973999857902527 |
| topics[2].domain.id | https://openalex.org/domains/4 |
| topics[2].domain.display_name | Health Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2746 |
| topics[2].subfield.display_name | Surgery |
| topics[2].display_name | Tissue Engineering and Regenerative Medicine |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C129536746 |
| concepts[0].level | 4 |
| concepts[0].score | 0.8699747920036316 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1141026 |
| concepts[0].display_name | Gastrulation |
| concepts[1].id | https://openalex.org/C75059768 |
| concepts[1].level | 5 |
| concepts[1].score | 0.7907817959785461 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q519516 |
| concepts[1].display_name | Epiblast |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.768548309803009 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C83330619 |
| concepts[3].level | 2 |
| concepts[3].score | 0.7515794038772583 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q21124872 |
| concepts[3].display_name | NODAL |
| concepts[4].id | https://openalex.org/C205583638 |
| concepts[4].level | 5 |
| concepts[4].score | 0.6676309704780579 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7046607 |
| concepts[4].display_name | Nodal signaling |
| concepts[5].id | https://openalex.org/C145103041 |
| concepts[5].level | 3 |
| concepts[5].score | 0.6652905941009521 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q1151519 |
| concepts[5].display_name | Embryonic stem cell |
| concepts[6].id | https://openalex.org/C95444343 |
| concepts[6].level | 1 |
| concepts[6].score | 0.6194948554039001 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[6].display_name | Cell biology |
| concepts[7].id | https://openalex.org/C137620995 |
| concepts[7].level | 3 |
| concepts[7].score | 0.6188843250274658 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q155769 |
| concepts[7].display_name | Wnt signaling pathway |
| concepts[8].id | https://openalex.org/C66518211 |
| concepts[8].level | 5 |
| concepts[8].score | 0.5716921091079712 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q247512 |
| concepts[8].display_name | Primitive streak |
| concepts[9].id | https://openalex.org/C127716648 |
| concepts[9].level | 3 |
| concepts[9].score | 0.5337041020393372 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q104053 |
| concepts[9].display_name | Phenotype |
| concepts[10].id | https://openalex.org/C2776817793 |
| concepts[10].level | 3 |
| concepts[10].score | 0.42448654770851135 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q6553369 |
| concepts[10].display_name | Lineage (genetic) |
| concepts[11].id | https://openalex.org/C196843134 |
| concepts[11].level | 2 |
| concepts[11].score | 0.42024126648902893 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q33196 |
| concepts[11].display_name | Embryo |
| concepts[12].id | https://openalex.org/C2777164004 |
| concepts[12].level | 4 |
| concepts[12].score | 0.41738489270210266 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q331858 |
| concepts[12].display_name | Endoderm |
| concepts[13].id | https://openalex.org/C54355233 |
| concepts[13].level | 1 |
| concepts[13].score | 0.33124226331710815 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[13].display_name | Genetics |
| concepts[14].id | https://openalex.org/C87073359 |
| concepts[14].level | 3 |
| concepts[14].score | 0.27413100004196167 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q33196 |
| concepts[14].display_name | Embryogenesis |
| concepts[15].id | https://openalex.org/C62478195 |
| concepts[15].level | 2 |
| concepts[15].score | 0.16160312294960022 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q828130 |
| concepts[15].display_name | Signal transduction |
| concepts[16].id | https://openalex.org/C104317684 |
| concepts[16].level | 2 |
| concepts[16].score | 0.11610835790634155 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[16].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/gastrulation |
| keywords[0].score | 0.8699747920036316 |
| keywords[0].display_name | Gastrulation |
| keywords[1].id | https://openalex.org/keywords/epiblast |
| keywords[1].score | 0.7907817959785461 |
| keywords[1].display_name | Epiblast |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.768548309803009 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/nodal |
| keywords[3].score | 0.7515794038772583 |
| keywords[3].display_name | NODAL |
| keywords[4].id | https://openalex.org/keywords/nodal-signaling |
| keywords[4].score | 0.6676309704780579 |
| keywords[4].display_name | Nodal signaling |
| keywords[5].id | https://openalex.org/keywords/embryonic-stem-cell |
| keywords[5].score | 0.6652905941009521 |
| keywords[5].display_name | Embryonic stem cell |
| keywords[6].id | https://openalex.org/keywords/cell-biology |
| keywords[6].score | 0.6194948554039001 |
| keywords[6].display_name | Cell biology |
| keywords[7].id | https://openalex.org/keywords/wnt-signaling-pathway |
| keywords[7].score | 0.6188843250274658 |
| keywords[7].display_name | Wnt signaling pathway |
| keywords[8].id | https://openalex.org/keywords/primitive-streak |
| keywords[8].score | 0.5716921091079712 |
| keywords[8].display_name | Primitive streak |
| keywords[9].id | https://openalex.org/keywords/phenotype |
| keywords[9].score | 0.5337041020393372 |
| keywords[9].display_name | Phenotype |
| keywords[10].id | https://openalex.org/keywords/lineage |
| keywords[10].score | 0.42448654770851135 |
| keywords[10].display_name | Lineage (genetic) |
| keywords[11].id | https://openalex.org/keywords/embryo |
| keywords[11].score | 0.42024126648902893 |
| keywords[11].display_name | Embryo |
| keywords[12].id | https://openalex.org/keywords/endoderm |
| keywords[12].score | 0.41738489270210266 |
| keywords[12].display_name | Endoderm |
| keywords[13].id | https://openalex.org/keywords/genetics |
| keywords[13].score | 0.33124226331710815 |
| keywords[13].display_name | Genetics |
| keywords[14].id | https://openalex.org/keywords/embryogenesis |
| keywords[14].score | 0.27413100004196167 |
| keywords[14].display_name | Embryogenesis |
| keywords[15].id | https://openalex.org/keywords/signal-transduction |
| keywords[15].score | 0.16160312294960022 |
| keywords[15].display_name | Signal transduction |
| keywords[16].id | https://openalex.org/keywords/gene |
| keywords[16].score | 0.11610835790634155 |
| keywords[16].display_name | Gene |
| language | en |
| locations[0].id | doi:10.1101/2020.11.30.404418 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2020.11.30.404418 |
| locations[1].id | pmh:oai:HAL:hal-03096917v1 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306402512 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | HAL (Le Centre pour la Communication Scientifique Directe) |
| locations[1].source.host_organization | https://openalex.org/I1294671590 |
| locations[1].source.host_organization_name | Centre National de la Recherche Scientifique |
| locations[1].source.host_organization_lineage | https://openalex.org/I1294671590 |
| locations[1].license | other-oa |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Preprints, Working Papers, ... |
| locations[1].license_id | https://openalex.org/licenses/other-oa |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | 2020 |
| locations[1].landing_page_url | https://hal.science/hal-03096917 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5051430111 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Jean-Louis Plouhinec |
| authorships[0].countries | FR |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210127056 |
| authorships[0].affiliations[0].raw_affiliation_string | Université de Paris, CNRS, Laboratoire Matière et système Complexes, Paris France |
| authorships[0].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[0].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[0].institutions[0].type | government |
| authorships[0].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[0].institutions[0].country_code | FR |
| authorships[0].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[0].institutions[1].id | https://openalex.org/I4210127056 |
| authorships[0].institutions[1].ror | https://ror.org/032w6q449 |
| authorships[0].institutions[1].type | facility |
| authorships[0].institutions[1].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210095849, https://openalex.org/I4210127056 |
| authorships[0].institutions[1].country_code | FR |
| authorships[0].institutions[1].display_name | Laboratoire Matière et Systèmes Complexes |
| authorships[0].institutions[2].id | https://openalex.org/I204730241 |
| authorships[0].institutions[2].ror | https://ror.org/05f82e368 |
| authorships[0].institutions[2].type | education |
| authorships[0].institutions[2].lineage | https://openalex.org/I204730241 |
| authorships[0].institutions[2].country_code | FR |
| authorships[0].institutions[2].display_name | Université Paris Cité |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jean-Louis Plouhinec |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Université de Paris, CNRS, Laboratoire Matière et système Complexes, Paris France |
| authorships[1].author.id | https://openalex.org/A5112584946 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Mathieu Vieira |
| authorships[1].countries | FR |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210113761 |
| authorships[1].affiliations[0].raw_affiliation_string | Université de Paris, CNRS, Institut Jacques Monod, Paris, France |
| authorships[1].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[1].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[1].institutions[0].type | government |
| authorships[1].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[1].institutions[0].country_code | FR |
| authorships[1].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[1].institutions[1].id | https://openalex.org/I4210113761 |
| authorships[1].institutions[1].ror | https://ror.org/02c5gc203 |
| authorships[1].institutions[1].type | facility |
| authorships[1].institutions[1].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I154526488, https://openalex.org/I204730241, https://openalex.org/I4210096427, https://openalex.org/I4210113761 |
| authorships[1].institutions[1].country_code | FR |
| authorships[1].institutions[1].display_name | Institut Jacques Monod |
| authorships[1].institutions[2].id | https://openalex.org/I204730241 |
| authorships[1].institutions[2].ror | https://ror.org/05f82e368 |
| authorships[1].institutions[2].type | education |
| authorships[1].institutions[2].lineage | https://openalex.org/I204730241 |
| authorships[1].institutions[2].country_code | FR |
| authorships[1].institutions[2].display_name | Université Paris Cité |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mathieu Vieira |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Université de Paris, CNRS, Institut Jacques Monod, Paris, France |
| authorships[2].author.id | https://openalex.org/A5059945915 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-9548-0240 |
| authorships[2].author.display_name | Gaël Simon |
| authorships[2].countries | FR |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210127056 |
| authorships[2].affiliations[0].raw_affiliation_string | Université de Paris, CNRS, Laboratoire Matière et système Complexes, Paris France |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210113761 |
| authorships[2].affiliations[1].raw_affiliation_string | Université de Paris, CNRS, Institut Jacques Monod, Paris, France |
| authorships[2].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[2].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[2].institutions[0].type | government |
| authorships[2].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[2].institutions[0].country_code | FR |
| authorships[2].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[2].institutions[1].id | https://openalex.org/I4210113761 |
| authorships[2].institutions[1].ror | https://ror.org/02c5gc203 |
| authorships[2].institutions[1].type | facility |
| authorships[2].institutions[1].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I154526488, https://openalex.org/I204730241, https://openalex.org/I4210096427, https://openalex.org/I4210113761 |
| authorships[2].institutions[1].country_code | FR |
| authorships[2].institutions[1].display_name | Institut Jacques Monod |
| authorships[2].institutions[2].id | https://openalex.org/I4210127056 |
| authorships[2].institutions[2].ror | https://ror.org/032w6q449 |
| authorships[2].institutions[2].type | facility |
| authorships[2].institutions[2].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210095849, https://openalex.org/I4210127056 |
| authorships[2].institutions[2].country_code | FR |
| authorships[2].institutions[2].display_name | Laboratoire Matière et Systèmes Complexes |
| authorships[2].institutions[3].id | https://openalex.org/I204730241 |
| authorships[2].institutions[3].ror | https://ror.org/05f82e368 |
| authorships[2].institutions[3].type | education |
| authorships[2].institutions[3].lineage | https://openalex.org/I204730241 |
| authorships[2].institutions[3].country_code | FR |
| authorships[2].institutions[3].display_name | Université Paris Cité |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Gaël Simon |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Université de Paris, CNRS, Institut Jacques Monod, Paris, France, Université de Paris, CNRS, Laboratoire Matière et système Complexes, Paris France |
| authorships[3].author.id | https://openalex.org/A5019695169 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-1212-4006 |
| authorships[3].author.display_name | Jérôme Collignon |
| authorships[3].countries | FR |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I1294671590, https://openalex.org/I204730241, https://openalex.org/I4210113761 |
| authorships[3].affiliations[0].raw_affiliation_string | Université de Paris, CNRS, Institut Jacques Monod, Paris, France |
| authorships[3].institutions[0].id | https://openalex.org/I1294671590 |
| authorships[3].institutions[0].ror | https://ror.org/02feahw73 |
| authorships[3].institutions[0].type | government |
| authorships[3].institutions[0].lineage | https://openalex.org/I1294671590 |
| authorships[3].institutions[0].country_code | FR |
| authorships[3].institutions[0].display_name | Centre National de la Recherche Scientifique |
| authorships[3].institutions[1].id | https://openalex.org/I4210113761 |
| authorships[3].institutions[1].ror | https://ror.org/02c5gc203 |
| authorships[3].institutions[1].type | facility |
| authorships[3].institutions[1].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I154526488, https://openalex.org/I204730241, https://openalex.org/I4210096427, https://openalex.org/I4210113761 |
| authorships[3].institutions[1].country_code | FR |
| authorships[3].institutions[1].display_name | Institut Jacques Monod |
| authorships[3].institutions[2].id | https://openalex.org/I204730241 |
| authorships[3].institutions[2].ror | https://ror.org/05f82e368 |
| authorships[3].institutions[2].type | education |
| authorships[3].institutions[2].lineage | https://openalex.org/I204730241 |
| authorships[3].institutions[2].country_code | FR |
| authorships[3].institutions[2].display_name | Université Paris Cité |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jérôme Collignon |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Université de Paris, CNRS, Institut Jacques Monod, Paris, France |
| authorships[4].author.id | https://openalex.org/A5078646816 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-8810-3298 |
| authorships[4].author.display_name | Benoît Sorre |
| authorships[4].countries | FR |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I4210160133, https://openalex.org/I80043 |
| authorships[4].affiliations[0].raw_affiliation_string | PCC - Laboratoire Physico-Chimie Curie [Institut Curie] (11 rue Pierre et Marie Curie 75005 Paris - France) |
| authorships[4].institutions[0].id | https://openalex.org/I80043 |
| authorships[4].institutions[0].ror | https://ror.org/04t0gwh46 |
| authorships[4].institutions[0].type | nonprofit |
| authorships[4].institutions[0].lineage | https://openalex.org/I80043 |
| authorships[4].institutions[0].country_code | FR |
| authorships[4].institutions[0].display_name | Institut Curie |
| authorships[4].institutions[1].id | https://openalex.org/I4210160133 |
| authorships[4].institutions[1].ror | https://ror.org/055ss7a31 |
| authorships[4].institutions[1].type | facility |
| authorships[4].institutions[1].lineage | https://openalex.org/I1294671590, https://openalex.org/I1294671590, https://openalex.org/I39804081, https://openalex.org/I4210096427, https://openalex.org/I4210160133, https://openalex.org/I80043 |
| authorships[4].institutions[1].country_code | FR |
| authorships[4].institutions[1].display_name | Physique des Cellules et Cancers |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Benoit Sorre |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | PCC - Laboratoire Physico-Chimie Curie [Institut Curie] (11 rue Pierre et Marie Curie 75005 Paris - France) |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Dissecting signalling hierarchies in the patterning of the mouse primitive streak using micro-patterned EpiLC colonies |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10505 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Pluripotent Stem Cells Research |
| related_works | https://openalex.org/W2031366529, https://openalex.org/W1971012578, https://openalex.org/W2152504473, https://openalex.org/W2007664084, https://openalex.org/W2063778808, https://openalex.org/W4283031190, https://openalex.org/W159742518, https://openalex.org/W2164563970, https://openalex.org/W2002842445, https://openalex.org/W2413083347 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2022 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1101/2020.11.30.404418 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2020.11.30.404418 |
| primary_location.id | doi:10.1101/2020.11.30.404418 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2020/11/30/2020.11.30.404418.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2020.11.30.404418 |
| publication_date | 2020-11-30 |
| publication_year | 2020 |
| referenced_works | https://openalex.org/W2070700342, https://openalex.org/W2169529166, https://openalex.org/W1964691304, https://openalex.org/W2125336256, https://openalex.org/W2069693786, https://openalex.org/W2142482111, https://openalex.org/W2044830131, https://openalex.org/W2158931884, https://openalex.org/W2117021242, https://openalex.org/W2041573813, https://openalex.org/W2052128821, https://openalex.org/W2417865499, https://openalex.org/W2147350988, https://openalex.org/W2050907040, https://openalex.org/W2132729210, https://openalex.org/W2535856472, https://openalex.org/W2024181148, https://openalex.org/W2100676040, https://openalex.org/W1973267391, https://openalex.org/W2048535898, https://openalex.org/W2023745283, https://openalex.org/W2161604785, https://openalex.org/W1977001077, https://openalex.org/W2951213089, https://openalex.org/W1982113242, https://openalex.org/W1541821200, https://openalex.org/W2941332545, https://openalex.org/W2045523681, https://openalex.org/W2786431878, https://openalex.org/W2129835280, https://openalex.org/W2142514820, https://openalex.org/W2061915791, https://openalex.org/W2113831186, https://openalex.org/W2306104084, https://openalex.org/W4244000493, https://openalex.org/W2093011529, https://openalex.org/W2912272542, https://openalex.org/W2009096862, https://openalex.org/W2027607261, https://openalex.org/W2091882935, https://openalex.org/W2148272837, https://openalex.org/W2583932040, https://openalex.org/W2055230542, https://openalex.org/W2076812266, https://openalex.org/W2090260381, https://openalex.org/W2164563970, https://openalex.org/W1998317358, https://openalex.org/W2053702504, https://openalex.org/W2107141327, https://openalex.org/W2016350172, https://openalex.org/W2127202275, https://openalex.org/W2071981231, https://openalex.org/W2063023858, https://openalex.org/W2082584450, https://openalex.org/W198385674, https://openalex.org/W2108133505, https://openalex.org/W2000041734, https://openalex.org/W1969141907, https://openalex.org/W2969998161, https://openalex.org/W2158470573 |
| referenced_works_count | 60 |
| abstract_inverted_index.a | 17, 74 |
| abstract_inverted_index.by | 127 |
| abstract_inverted_index.in | 115, 118 |
| abstract_inverted_index.is | 14, 65, 100 |
| abstract_inverted_index.it | 67 |
| abstract_inverted_index.of | 9, 32, 53, 84, 129, 139 |
| abstract_inverted_index.on | 16, 45 |
| abstract_inverted_index.or | 131 |
| abstract_inverted_index.to | 36, 77, 92, 96, 102, 143 |
| abstract_inverted_index.us | 91 |
| abstract_inverted_index.we | 60 |
| abstract_inverted_index.BMP | 23, 64, 97 |
| abstract_inverted_index.Our | 134 |
| abstract_inverted_index.The | 107 |
| abstract_inverted_index.WNT | 71 |
| abstract_inverted_index.and | 24, 49, 70, 72 |
| abstract_inverted_index.are | 124 |
| abstract_inverted_index.the | 7, 10, 21, 46, 50, 78, 82, 104, 137 |
| abstract_inverted_index.two | 85 |
| abstract_inverted_index.use | 83 |
| abstract_inverted_index.Like | 56 |
| abstract_inverted_index.WNT, | 22 |
| abstract_inverted_index.cell | 88 |
| abstract_inverted_index.fact | 108 |
| abstract_inverted_index.have | 4 |
| abstract_inverted_index.more | 113 |
| abstract_inverted_index.show | 61, 93 |
| abstract_inverted_index.than | 117 |
| abstract_inverted_index.that | 6, 62, 94, 109, 121 |
| abstract_inverted_index.this | 37 |
| abstract_inverted_index.vivo | 119 |
| abstract_inverted_index.were | 112 |
| abstract_inverted_index.when | 63 |
| abstract_inverted_index.work | 135 |
| abstract_inverted_index.Cells | 57 |
| abstract_inverted_index.EpiLC | 141 |
| abstract_inverted_index.Here, | 41 |
| abstract_inverted_index.NODAL | 25, 69, 99 |
| abstract_inverted_index.Nodal | 86 |
| abstract_inverted_index.lines | 89 |
| abstract_inverted_index.mouse | 12 |
| abstract_inverted_index.prior | 95 |
| abstract_inverted_index.their | 33, 43 |
| abstract_inverted_index.vitro | 116 |
| abstract_inverted_index.where | 20 |
| abstract_inverted_index.Still, | 29 |
| abstract_inverted_index.embryo | 13 |
| abstract_inverted_index.impact | 44 |
| abstract_inverted_index.mutant | 87, 110 |
| abstract_inverted_index.remain | 39 |
| abstract_inverted_index.severe | 114 |
| abstract_inverted_index.(EpiLC) | 58 |
| abstract_inverted_index.action, | 98 |
| abstract_inverted_index.allowed | 90 |
| abstract_inverted_index.aspects | 31 |
| abstract_inverted_index.imposes | 73 |
| abstract_inverted_index.ligands | 128 |
| abstract_inverted_index.network | 19 |
| abstract_inverted_index.origin. | 133 |
| abstract_inverted_index.process | 38 |
| abstract_inverted_index.rescued | 126 |
| abstract_inverted_index.spatial | 47 |
| abstract_inverted_index.studies | 3 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Epiblast | 55 |
| abstract_inverted_index.However, | 81 |
| abstract_inverted_index.colonies | 79, 142 |
| abstract_inverted_index.lineage. | 106 |
| abstract_inverted_index.maternal | 132 |
| abstract_inverted_index.pathways | 27 |
| abstract_inverted_index.present, | 66 |
| abstract_inverted_index.required | 101 |
| abstract_inverted_index.studying | 42 |
| abstract_inverted_index.suggests | 120 |
| abstract_inverted_index.unclear. | 40 |
| abstract_inverted_index.Molecular | 1 |
| abstract_inverted_index.character | 76 |
| abstract_inverted_index.colonies, | 59 |
| abstract_inverted_index.dependent | 15 |
| abstract_inverted_index.dominates | 68 |
| abstract_inverted_index.embryonic | 122 |
| abstract_inverted_index.establish | 103 |
| abstract_inverted_index.important | 30 |
| abstract_inverted_index.partially | 125 |
| abstract_inverted_index.posterior | 75 |
| abstract_inverted_index.cooperate. | 28 |
| abstract_inverted_index.embryology | 2 |
| abstract_inverted_index.patterning | 8 |
| abstract_inverted_index.phenotypes | 111, 123 |
| abstract_inverted_index.regulatory | 18 |
| abstract_inverted_index.respective | 34 |
| abstract_inverted_index.signalling | 26 |
| abstract_inverted_index.approaches. | 145 |
| abstract_inverted_index.established | 5 |
| abstract_inverted_index.demonstrates | 136 |
| abstract_inverted_index.organization | 48 |
| abstract_inverted_index.trajectories | 52 |
| abstract_inverted_index.contributions | 35 |
| abstract_inverted_index.developmental | 51 |
| abstract_inverted_index.embryological | 144 |
| abstract_inverted_index.mesendodermal | 105 |
| abstract_inverted_index.gastrula-stage | 11 |
| abstract_inverted_index.complementarity | 138 |
| abstract_inverted_index.extra-embryonic | 130 |
| abstract_inverted_index.micro-patterned | 54, 140 |
| abstract_inverted_index.differentiation. | 80 |
| cited_by_percentile_year.max | 94 |
| cited_by_percentile_year.min | 89 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 5 |
| citation_normalized_percentile.value | 0.47127143 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |