Divergent Cl− and H+ pathways underlie transport coupling and gating in CLC exchangers and channels Article Swipe
 YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1101/753954
YOU?
·
· 2019
· Open Access
·
· DOI: https://doi.org/10.1101/753954
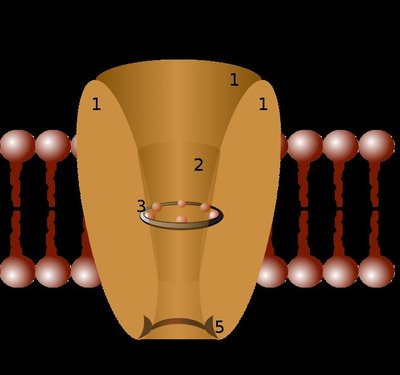
The CLC family of anion transporting proteins is comprised of secondary active H + -coupled exchangers and of Cl − channels. Both functional subtypes play key roles in human physiology, and mutations causing their dysfunction lead to numerous genetic disorders. Current models suggest that the CLC exchangers do not utilize a classical ‘ping-pong’ mechanism of antiport, where the transporter sequentially interacts with one substrate at a time. Rather, in the CLC exchangers both substrates bind and translocate simultaneously while moving through partially congruent pathways. How ions of opposite electrical charge bypass each other while moving in opposite directions through a shared permeation pathway remains unknown. Here, we use MD simulations in combination with biochemical and electrophysiological measurements to identify a pair of highly conserved phenylalanine residues that form an aromatic pathway, separate from the Cl − pore, whose dynamic rearrangements enable H + movement. Mutations of these aromatic residues impair H + transport and voltage-dependent gating in the CLC exchangers. Remarkably, the role of the aromatic pathway is evolutionarily conserved in CLC channels. Using atomic-scale mutagenesis we show that the electrostatic properties and conformational flexibility of these aromatic residues are essential determinants of channel gating. Our results suggest that Cl − and H + move through physically distinct and evolutionarily conserved routes through the CLC channels and transporters. We propose a unifying mechanism that describes the gating mechanism of CLC exchangers and channels.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/753954
- https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdf
- OA Status
- green
- References
- 60
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W2971858961
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2971858961Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/753954Digital Object Identifier
- Title
-
Divergent Cl− and H+ pathways underlie transport coupling and gating in CLC exchangers and channelsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2019Year of publication
- Publication date
-
2019-09-05Full publication date if available
- Authors
-
Lilia Leisle, Yanyan Xu, Eva Fortea, Jason D. Galpin, Malvin Vien, Christopher A. Ahern, Alessio Accardi, Simon BernècheList of authors in order
- Landing page
-
https://doi.org/10.1101/753954Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdfDirect OA link when available
- Concepts
-
Gating, Chemistry, Antiporter, Biophysics, Transporter, Ion channel, Ion transporter, Coupling (piping), Stereochemistry, Biochemistry, Membrane, Biology, Gene, Materials science, Receptor, MetallurgyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
60Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2971858961 |
|---|---|
| doi | https://doi.org/10.1101/753954 |
| ids.doi | https://doi.org/10.1101/753954 |
| ids.mag | 2971858961 |
| ids.openalex | https://openalex.org/W2971858961 |
| fwci | 0.0 |
| type | preprint |
| title | Divergent Cl− and H+ pathways underlie transport coupling and gating in CLC exchangers and channels |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10493 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Ion channel regulation and function |
| topics[1].id | https://openalex.org/T10077 |
| topics[1].field.id | https://openalex.org/fields/28 |
| topics[1].field.display_name | Neuroscience |
| topics[1].score | 0.9991000294685364 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2804 |
| topics[1].subfield.display_name | Cellular and Molecular Neuroscience |
| topics[1].display_name | Neuroscience and Neuropharmacology Research |
| topics[2].id | https://openalex.org/T11434 |
| topics[2].field.id | https://openalex.org/fields/16 |
| topics[2].field.display_name | Chemistry |
| topics[2].score | 0.9972000122070312 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1603 |
| topics[2].subfield.display_name | Electrochemistry |
| topics[2].display_name | Electrochemical Analysis and Applications |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C194544171 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8682719469070435 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21105679 |
| concepts[0].display_name | Gating |
| concepts[1].id | https://openalex.org/C185592680 |
| concepts[1].level | 0 |
| concepts[1].score | 0.6822508573532104 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[1].display_name | Chemistry |
| concepts[2].id | https://openalex.org/C57181701 |
| concepts[2].level | 3 |
| concepts[2].score | 0.6416943073272705 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2853339 |
| concepts[2].display_name | Antiporter |
| concepts[3].id | https://openalex.org/C12554922 |
| concepts[3].level | 1 |
| concepts[3].score | 0.6324710249900818 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7100 |
| concepts[3].display_name | Biophysics |
| concepts[4].id | https://openalex.org/C149011108 |
| concepts[4].level | 3 |
| concepts[4].score | 0.4573172926902771 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q652985 |
| concepts[4].display_name | Transporter |
| concepts[5].id | https://openalex.org/C50254741 |
| concepts[5].level | 3 |
| concepts[5].score | 0.4535256028175354 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q62536 |
| concepts[5].display_name | Ion channel |
| concepts[6].id | https://openalex.org/C99975950 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4479590356349945 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q911011 |
| concepts[6].display_name | Ion transporter |
| concepts[7].id | https://openalex.org/C131584629 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4207690358161926 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q4308705 |
| concepts[7].display_name | Coupling (piping) |
| concepts[8].id | https://openalex.org/C71240020 |
| concepts[8].level | 1 |
| concepts[8].score | 0.34278810024261475 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q186011 |
| concepts[8].display_name | Stereochemistry |
| concepts[9].id | https://openalex.org/C55493867 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3197535276412964 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[9].display_name | Biochemistry |
| concepts[10].id | https://openalex.org/C41625074 |
| concepts[10].level | 2 |
| concepts[10].score | 0.2832362651824951 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[10].display_name | Membrane |
| concepts[11].id | https://openalex.org/C86803240 |
| concepts[11].level | 0 |
| concepts[11].score | 0.14577332139015198 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[11].display_name | Biology |
| concepts[12].id | https://openalex.org/C104317684 |
| concepts[12].level | 2 |
| concepts[12].score | 0.1116115152835846 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[12].display_name | Gene |
| concepts[13].id | https://openalex.org/C192562407 |
| concepts[13].level | 0 |
| concepts[13].score | 0.07931888103485107 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q228736 |
| concepts[13].display_name | Materials science |
| concepts[14].id | https://openalex.org/C170493617 |
| concepts[14].level | 2 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[14].display_name | Receptor |
| concepts[15].id | https://openalex.org/C191897082 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q11467 |
| concepts[15].display_name | Metallurgy |
| keywords[0].id | https://openalex.org/keywords/gating |
| keywords[0].score | 0.8682719469070435 |
| keywords[0].display_name | Gating |
| keywords[1].id | https://openalex.org/keywords/chemistry |
| keywords[1].score | 0.6822508573532104 |
| keywords[1].display_name | Chemistry |
| keywords[2].id | https://openalex.org/keywords/antiporter |
| keywords[2].score | 0.6416943073272705 |
| keywords[2].display_name | Antiporter |
| keywords[3].id | https://openalex.org/keywords/biophysics |
| keywords[3].score | 0.6324710249900818 |
| keywords[3].display_name | Biophysics |
| keywords[4].id | https://openalex.org/keywords/transporter |
| keywords[4].score | 0.4573172926902771 |
| keywords[4].display_name | Transporter |
| keywords[5].id | https://openalex.org/keywords/ion-channel |
| keywords[5].score | 0.4535256028175354 |
| keywords[5].display_name | Ion channel |
| keywords[6].id | https://openalex.org/keywords/ion-transporter |
| keywords[6].score | 0.4479590356349945 |
| keywords[6].display_name | Ion transporter |
| keywords[7].id | https://openalex.org/keywords/coupling |
| keywords[7].score | 0.4207690358161926 |
| keywords[7].display_name | Coupling (piping) |
| keywords[8].id | https://openalex.org/keywords/stereochemistry |
| keywords[8].score | 0.34278810024261475 |
| keywords[8].display_name | Stereochemistry |
| keywords[9].id | https://openalex.org/keywords/biochemistry |
| keywords[9].score | 0.3197535276412964 |
| keywords[9].display_name | Biochemistry |
| keywords[10].id | https://openalex.org/keywords/membrane |
| keywords[10].score | 0.2832362651824951 |
| keywords[10].display_name | Membrane |
| keywords[11].id | https://openalex.org/keywords/biology |
| keywords[11].score | 0.14577332139015198 |
| keywords[11].display_name | Biology |
| keywords[12].id | https://openalex.org/keywords/gene |
| keywords[12].score | 0.1116115152835846 |
| keywords[12].display_name | Gene |
| keywords[13].id | https://openalex.org/keywords/materials-science |
| keywords[13].score | 0.07931888103485107 |
| keywords[13].display_name | Materials science |
| language | en |
| locations[0].id | doi:10.1101/753954 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/753954 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5001412216 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-3987-4269 |
| authorships[0].author.display_name | Lilia Leisle |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I205783295 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Anesthesiology, Weill Cornell Medical College |
| authorships[0].institutions[0].id | https://openalex.org/I205783295 |
| authorships[0].institutions[0].ror | https://ror.org/05bnh6r87 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I205783295 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Cornell University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Lilia Leisle |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Anesthesiology, Weill Cornell Medical College |
| authorships[1].author.id | https://openalex.org/A5023103908 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-8152-7423 |
| authorships[1].author.display_name | Yanyan Xu |
| authorships[1].countries | CH |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I1850255 |
| authorships[1].affiliations[0].raw_affiliation_string | Biozentrum, University of Basel 50/70 Klingelbergstrasse |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I12708293, https://openalex.org/I1850255 |
| authorships[1].affiliations[1].raw_affiliation_string | SIB Swiss Institute of Bioinformatics, University of Basel 50/70 Klingelbergstrasse |
| authorships[1].institutions[0].id | https://openalex.org/I12708293 |
| authorships[1].institutions[0].ror | https://ror.org/002n09z45 |
| authorships[1].institutions[0].type | nonprofit |
| authorships[1].institutions[0].lineage | https://openalex.org/I12708293 |
| authorships[1].institutions[0].country_code | CH |
| authorships[1].institutions[0].display_name | SIB Swiss Institute of Bioinformatics |
| authorships[1].institutions[1].id | https://openalex.org/I1850255 |
| authorships[1].institutions[1].ror | https://ror.org/02s6k3f65 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I1850255 |
| authorships[1].institutions[1].country_code | CH |
| authorships[1].institutions[1].display_name | University of Basel |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Yanyan Xu |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Biozentrum, University of Basel 50/70 Klingelbergstrasse, SIB Swiss Institute of Bioinformatics, University of Basel 50/70 Klingelbergstrasse |
| authorships[2].author.id | https://openalex.org/A5030352462 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-4328-8940 |
| authorships[2].author.display_name | Eva Fortea |
| authorships[2].countries | US |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I205783295 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Physiology and Biophysics, Weill Cornell Medical College |
| authorships[2].institutions[0].id | https://openalex.org/I205783295 |
| authorships[2].institutions[0].ror | https://ror.org/05bnh6r87 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I205783295 |
| authorships[2].institutions[0].country_code | US |
| authorships[2].institutions[0].display_name | Cornell University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Eva Fortea |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Physiology and Biophysics, Weill Cornell Medical College |
| authorships[3].author.id | https://openalex.org/A5006834297 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-1923-5312 |
| authorships[3].author.display_name | Jason D. Galpin |
| authorships[3].countries | US |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I126307644 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Molecular Physiology and Biophysics, University of Iowa |
| authorships[3].institutions[0].id | https://openalex.org/I126307644 |
| authorships[3].institutions[0].ror | https://ror.org/036jqmy94 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I126307644 |
| authorships[3].institutions[0].country_code | US |
| authorships[3].institutions[0].display_name | University of Iowa |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jason Galpin |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Molecular Physiology and Biophysics, University of Iowa |
| authorships[4].author.id | https://openalex.org/A5035992397 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Malvin Vien |
| authorships[4].countries | US |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I205783295 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Anesthesiology, Weill Cornell Medical College |
| authorships[4].institutions[0].id | https://openalex.org/I205783295 |
| authorships[4].institutions[0].ror | https://ror.org/05bnh6r87 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I205783295 |
| authorships[4].institutions[0].country_code | US |
| authorships[4].institutions[0].display_name | Cornell University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Malvin Vien |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Anesthesiology, Weill Cornell Medical College |
| authorships[5].author.id | https://openalex.org/A5038268798 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-7975-2744 |
| authorships[5].author.display_name | Christopher A. Ahern |
| authorships[5].countries | US |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I126307644 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Molecular Physiology and Biophysics, University of Iowa |
| authorships[5].institutions[0].id | https://openalex.org/I126307644 |
| authorships[5].institutions[0].ror | https://ror.org/036jqmy94 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I126307644 |
| authorships[5].institutions[0].country_code | US |
| authorships[5].institutions[0].display_name | University of Iowa |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Christopher A. Ahern |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Molecular Physiology and Biophysics, University of Iowa |
| authorships[6].author.id | https://openalex.org/A5040398803 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-6584-0102 |
| authorships[6].author.display_name | Alessio Accardi |
| authorships[6].countries | US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I205783295 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Biochemistry, Weill Cornell Medical College |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I205783295 |
| authorships[6].affiliations[1].raw_affiliation_string | Department of Anesthesiology, Weill Cornell Medical College |
| authorships[6].affiliations[2].institution_ids | https://openalex.org/I205783295 |
| authorships[6].affiliations[2].raw_affiliation_string | Department of Physiology and Biophysics, Weill Cornell Medical College |
| authorships[6].institutions[0].id | https://openalex.org/I205783295 |
| authorships[6].institutions[0].ror | https://ror.org/05bnh6r87 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I205783295 |
| authorships[6].institutions[0].country_code | US |
| authorships[6].institutions[0].display_name | Cornell University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Alessio Accardi |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Anesthesiology, Weill Cornell Medical College, Department of Biochemistry, Weill Cornell Medical College, Department of Physiology and Biophysics, Weill Cornell Medical College |
| authorships[7].author.id | https://openalex.org/A5071177698 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-6274-4094 |
| authorships[7].author.display_name | Simon Bernèche |
| authorships[7].countries | CH |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I12708293, https://openalex.org/I1850255 |
| authorships[7].affiliations[0].raw_affiliation_string | SIB Swiss Institute of Bioinformatics, University of Basel 50/70 Klingelbergstrasse |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I1850255 |
| authorships[7].affiliations[1].raw_affiliation_string | Biozentrum, University of Basel 50/70 Klingelbergstrasse |
| authorships[7].institutions[0].id | https://openalex.org/I12708293 |
| authorships[7].institutions[0].ror | https://ror.org/002n09z45 |
| authorships[7].institutions[0].type | nonprofit |
| authorships[7].institutions[0].lineage | https://openalex.org/I12708293 |
| authorships[7].institutions[0].country_code | CH |
| authorships[7].institutions[0].display_name | SIB Swiss Institute of Bioinformatics |
| authorships[7].institutions[1].id | https://openalex.org/I1850255 |
| authorships[7].institutions[1].ror | https://ror.org/02s6k3f65 |
| authorships[7].institutions[1].type | education |
| authorships[7].institutions[1].lineage | https://openalex.org/I1850255 |
| authorships[7].institutions[1].country_code | CH |
| authorships[7].institutions[1].display_name | University of Basel |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Simon Bernèche |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Biozentrum, University of Basel 50/70 Klingelbergstrasse, SIB Swiss Institute of Bioinformatics, University of Basel 50/70 Klingelbergstrasse |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Divergent Cl− and H+ pathways underlie transport coupling and gating in CLC exchangers and channels |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10493 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Ion channel regulation and function |
| related_works | https://openalex.org/W2087256484, https://openalex.org/W1966343975, https://openalex.org/W2232251872, https://openalex.org/W2013146219, https://openalex.org/W2225689529, https://openalex.org/W2996554726, https://openalex.org/W2043128249, https://openalex.org/W2111309902, https://openalex.org/W3192752836, https://openalex.org/W4292849647 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/753954 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/753954 |
| primary_location.id | doi:10.1101/753954 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2019/09/05/753954.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/753954 |
| publication_date | 2019-09-05 |
| publication_year | 2019 |
| referenced_works | https://openalex.org/W1983411645, https://openalex.org/W1994088425, https://openalex.org/W2120242708, https://openalex.org/W1965620778, https://openalex.org/W1509035510, https://openalex.org/W2105559183, https://openalex.org/W1948653993, https://openalex.org/W1982831381, https://openalex.org/W2060872117, https://openalex.org/W2803610788, https://openalex.org/W2132262459, https://openalex.org/W2127059741, https://openalex.org/W2164797647, https://openalex.org/W2005501525, https://openalex.org/W2035687084, https://openalex.org/W2143082492, https://openalex.org/W1980130504, https://openalex.org/W2146201767, https://openalex.org/W2000707162, https://openalex.org/W2055980566, https://openalex.org/W2795304024, https://openalex.org/W2069179913, https://openalex.org/W2105095499, https://openalex.org/W2805997389, https://openalex.org/W2021520922, https://openalex.org/W2281086844, https://openalex.org/W2151176467, https://openalex.org/W2112154906, https://openalex.org/W2808791900, https://openalex.org/W2103104037, https://openalex.org/W2565335750, https://openalex.org/W2067468321, https://openalex.org/W2048457888, https://openalex.org/W2090933371, https://openalex.org/W1995332683, https://openalex.org/W2039708016, https://openalex.org/W1970674037, https://openalex.org/W2067219399, https://openalex.org/W1987018129, https://openalex.org/W1496902833, https://openalex.org/W2563306975, https://openalex.org/W2806551698, https://openalex.org/W2144865316, https://openalex.org/W2141868802, https://openalex.org/W2124376775, https://openalex.org/W2055750879, https://openalex.org/W1990361339, https://openalex.org/W2052351570, https://openalex.org/W2562851776, https://openalex.org/W2150986640, https://openalex.org/W2122901539, https://openalex.org/W2594185052, https://openalex.org/W2941604165, https://openalex.org/W2002561835, https://openalex.org/W1990771438, https://openalex.org/W2030338986, https://openalex.org/W2072830920, https://openalex.org/W2146941780, https://openalex.org/W4285719527, https://openalex.org/W2506269565 |
| referenced_works_count | 60 |
| abstract_inverted_index.+ | 14, 143, 152, 204 |
| abstract_inverted_index.H | 13, 142, 151, 203 |
| abstract_inverted_index.a | 51, 66, 100, 120, 221 |
| abstract_inverted_index.Cl | 19, 135, 200 |
| abstract_inverted_index.MD | 109 |
| abstract_inverted_index.We | 219 |
| abstract_inverted_index.an | 129 |
| abstract_inverted_index.at | 65 |
| abstract_inverted_index.do | 48 |
| abstract_inverted_index.in | 28, 69, 96, 111, 157, 171 |
| abstract_inverted_index.is | 8, 168 |
| abstract_inverted_index.of | 4, 10, 18, 55, 87, 122, 146, 164, 186, 193, 229 |
| abstract_inverted_index.to | 37, 118 |
| abstract_inverted_index.we | 107, 177 |
| abstract_inverted_index.CLC | 2, 46, 71, 159, 172, 215, 230 |
| abstract_inverted_index.How | 85 |
| abstract_inverted_index.Our | 196 |
| abstract_inverted_index.The | 1 |
| abstract_inverted_index.and | 17, 31, 76, 115, 154, 183, 202, 209, 217, 232 |
| abstract_inverted_index.are | 190 |
| abstract_inverted_index.key | 26 |
| abstract_inverted_index.not | 49 |
| abstract_inverted_index.one | 63 |
| abstract_inverted_index.the | 45, 58, 70, 134, 158, 162, 165, 180, 214, 226 |
| abstract_inverted_index.use | 108 |
| abstract_inverted_index.− | 20, 136, 201 |
| abstract_inverted_index.Both | 22 |
| abstract_inverted_index.bind | 75 |
| abstract_inverted_index.both | 73 |
| abstract_inverted_index.each | 92 |
| abstract_inverted_index.form | 128 |
| abstract_inverted_index.from | 133 |
| abstract_inverted_index.ions | 86 |
| abstract_inverted_index.lead | 36 |
| abstract_inverted_index.move | 205 |
| abstract_inverted_index.pair | 121 |
| abstract_inverted_index.play | 25 |
| abstract_inverted_index.role | 163 |
| abstract_inverted_index.show | 178 |
| abstract_inverted_index.that | 44, 127, 179, 199, 224 |
| abstract_inverted_index.with | 62, 113 |
| abstract_inverted_index.Here, | 106 |
| abstract_inverted_index.Using | 174 |
| abstract_inverted_index.anion | 5 |
| abstract_inverted_index.human | 29 |
| abstract_inverted_index.other | 93 |
| abstract_inverted_index.pore, | 137 |
| abstract_inverted_index.roles | 27 |
| abstract_inverted_index.their | 34 |
| abstract_inverted_index.these | 147, 187 |
| abstract_inverted_index.time. | 67 |
| abstract_inverted_index.where | 57 |
| abstract_inverted_index.while | 79, 94 |
| abstract_inverted_index.whose | 138 |
| abstract_inverted_index.active | 12 |
| abstract_inverted_index.bypass | 91 |
| abstract_inverted_index.charge | 90 |
| abstract_inverted_index.enable | 141 |
| abstract_inverted_index.family | 3 |
| abstract_inverted_index.gating | 156, 227 |
| abstract_inverted_index.highly | 123 |
| abstract_inverted_index.impair | 150 |
| abstract_inverted_index.models | 42 |
| abstract_inverted_index.moving | 80, 95 |
| abstract_inverted_index.routes | 212 |
| abstract_inverted_index.shared | 101 |
| abstract_inverted_index.Current | 41 |
| abstract_inverted_index.Rather, | 68 |
| abstract_inverted_index.causing | 33 |
| abstract_inverted_index.channel | 194 |
| abstract_inverted_index.dynamic | 139 |
| abstract_inverted_index.gating. | 195 |
| abstract_inverted_index.genetic | 39 |
| abstract_inverted_index.pathway | 103, 167 |
| abstract_inverted_index.propose | 220 |
| abstract_inverted_index.remains | 104 |
| abstract_inverted_index.results | 197 |
| abstract_inverted_index.suggest | 43, 198 |
| abstract_inverted_index.through | 81, 99, 206, 213 |
| abstract_inverted_index.utilize | 50 |
| abstract_inverted_index.-coupled | 15 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.aromatic | 130, 148, 166, 188 |
| abstract_inverted_index.channels | 216 |
| abstract_inverted_index.distinct | 208 |
| abstract_inverted_index.identify | 119 |
| abstract_inverted_index.numerous | 38 |
| abstract_inverted_index.opposite | 88, 97 |
| abstract_inverted_index.pathway, | 131 |
| abstract_inverted_index.proteins | 7 |
| abstract_inverted_index.residues | 126, 149, 189 |
| abstract_inverted_index.separate | 132 |
| abstract_inverted_index.subtypes | 24 |
| abstract_inverted_index.unifying | 222 |
| abstract_inverted_index.unknown. | 105 |
| abstract_inverted_index.Mutations | 145 |
| abstract_inverted_index.antiport, | 56 |
| abstract_inverted_index.channels. | 21, 173, 233 |
| abstract_inverted_index.classical | 52 |
| abstract_inverted_index.comprised | 9 |
| abstract_inverted_index.congruent | 83 |
| abstract_inverted_index.conserved | 124, 170, 211 |
| abstract_inverted_index.describes | 225 |
| abstract_inverted_index.essential | 191 |
| abstract_inverted_index.interacts | 61 |
| abstract_inverted_index.mechanism | 54, 223, 228 |
| abstract_inverted_index.movement. | 144 |
| abstract_inverted_index.mutations | 32 |
| abstract_inverted_index.partially | 82 |
| abstract_inverted_index.pathways. | 84 |
| abstract_inverted_index.secondary | 11 |
| abstract_inverted_index.substrate | 64 |
| abstract_inverted_index.transport | 153 |
| abstract_inverted_index.directions | 98 |
| abstract_inverted_index.disorders. | 40 |
| abstract_inverted_index.electrical | 89 |
| abstract_inverted_index.exchangers | 16, 47, 72, 231 |
| abstract_inverted_index.functional | 23 |
| abstract_inverted_index.permeation | 102 |
| abstract_inverted_index.physically | 207 |
| abstract_inverted_index.properties | 182 |
| abstract_inverted_index.substrates | 74 |
| abstract_inverted_index.Remarkably, | 161 |
| abstract_inverted_index.biochemical | 114 |
| abstract_inverted_index.combination | 112 |
| abstract_inverted_index.dysfunction | 35 |
| abstract_inverted_index.exchangers. | 160 |
| abstract_inverted_index.flexibility | 185 |
| abstract_inverted_index.mutagenesis | 176 |
| abstract_inverted_index.physiology, | 30 |
| abstract_inverted_index.simulations | 110 |
| abstract_inverted_index.translocate | 77 |
| abstract_inverted_index.transporter | 59 |
| abstract_inverted_index.atomic-scale | 175 |
| abstract_inverted_index.determinants | 192 |
| abstract_inverted_index.measurements | 117 |
| abstract_inverted_index.sequentially | 60 |
| abstract_inverted_index.transporting | 6 |
| abstract_inverted_index.electrostatic | 181 |
| abstract_inverted_index.phenylalanine | 125 |
| abstract_inverted_index.transporters. | 218 |
| abstract_inverted_index.conformational | 184 |
| abstract_inverted_index.evolutionarily | 169, 210 |
| abstract_inverted_index.rearrangements | 140 |
| abstract_inverted_index.simultaneously | 78 |
| abstract_inverted_index.‘ping-pong’ | 53 |
| abstract_inverted_index.voltage-dependent | 155 |
| abstract_inverted_index.electrophysiological | 116 |
| cited_by_percentile_year | |
| countries_distinct_count | 2 |
| institutions_distinct_count | 8 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/6 |
| sustainable_development_goals[0].score | 0.41999998688697815 |
| sustainable_development_goals[0].display_name | Clean water and sanitation |
| citation_normalized_percentile.value | 0.08364042 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |