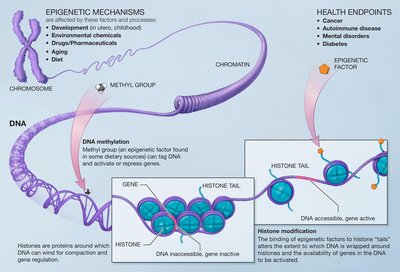
DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contexts Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1186/s13059-024-03218-6
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1186/s13059-024-03218-6
Background DNA methylation is an important epigenetic modification which has numerous roles in modulating genome function. Its levels are spatially correlated across the genome, typically high in repressed regions but low in transcription factor (TF) binding sites and active regulatory regions. However, the mechanisms establishing genome-wide and TF binding site methylation patterns are still unclear. Results Here we use a comparative approach to investigate the association of DNA methylation to TF binding evolution in mammals. Specifically, we experimentally profile DNA methylation and combine this with published occupancy profiles of five distinct TFs (CTCF, CEBPA, HNF4A, ONECUT1, FOXA1) in the liver of five mammalian species (human, macaque, mouse, rat, dog). TF binding sites are lowly methylated, but they often also have intermediate methylation levels. Furthermore, biding sites are influenced by the methylation status of CpGs in their wider binding regions even when CpGs are absent from the core binding motif. Employing a classification and clustering approach, we extract distinct and species-conserved patterns of DNA methylation levels at TF binding regions. CEBPA, HNF4A, ONECUT1, and FOXA1 share the same methylation patterns, while CTCF's differ. These patterns characterize alternative functions and chromatin landscapes of TF-bound regions. Leveraging our phylogenetic framework, we find DNA methylation gain upon evolutionary loss of TF occupancy, indicating coordinated evolution. Furthermore, each methylation pattern has its own evolutionary trajectory reflecting its genomic contexts. Conclusions Our epigenomic analyses indicate a role for DNA methylation in TF binding changes across species including that specific DNA methylation profiles characterize TF binding and are associated with their regulatory activity, chromatin contexts, and evolutionary trajectories.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1186/s13059-024-03218-6
- https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6
- OA Status
- gold
- Cited By
- 21
- References
- 66
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4399388201
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4399388201Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1186/s13059-024-03218-6Digital Object Identifier
- Title
-
DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contextsWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-06-06Full publication date if available
- Authors
-
Martina Rimoldi, Ning Wang, Jilin Zhang, Diego Villar, Duncan T. Odom, Jussi Taipale, Paul Flicek, Maša RollerList of authors in order
- Landing page
-
https://doi.org/10.1186/s13059-024-03218-6Publisher landing page
- PDF URL
-
https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6Direct link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6Direct OA link when available
- Concepts
-
DNA methylation, Biology, DNA binding site, Epigenomics, Genetics, Methylation, Epigenetics, RNA-Directed DNA Methylation, CTCF, Transcription factor, Chromatin, DNA, Computational biology, Gene, Promoter, Enhancer, Gene expressionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
21Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 15, 2024: 3, 2023: 3Per-year citation counts (last 5 years)
- References (count)
-
66Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4399388201 |
|---|---|
| doi | https://doi.org/10.1186/s13059-024-03218-6 |
| ids.doi | https://doi.org/10.1186/s13059-024-03218-6 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/38844976 |
| ids.openalex | https://openalex.org/W4399388201 |
| fwci | 8.64445005 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D000818 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Animals |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D019175 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | DNA Methylation |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D001665 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Binding Sites |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D006801 |
| mesh[3].is_major_topic | False |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Humans |
| mesh[4].qualifier_ui | Q000378 |
| mesh[4].descriptor_ui | D014157 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | metabolism |
| mesh[4].descriptor_name | Transcription Factors |
| mesh[5].qualifier_ui | Q000235 |
| mesh[5].descriptor_ui | D014157 |
| mesh[5].is_major_topic | True |
| mesh[5].qualifier_name | genetics |
| mesh[5].descriptor_name | Transcription Factors |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D019143 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Evolution, Molecular |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D051379 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Mice |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D051381 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Rats |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D018899 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | CpG Islands |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D004285 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Dogs |
| mesh[11].qualifier_ui | Q000378 |
| mesh[11].descriptor_ui | D051541 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | metabolism |
| mesh[11].descriptor_name | Hepatocyte Nuclear Factor 3-alpha |
| mesh[12].qualifier_ui | Q000235 |
| mesh[12].descriptor_ui | D051541 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | genetics |
| mesh[12].descriptor_name | Hepatocyte Nuclear Factor 3-alpha |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D011485 |
| mesh[13].is_major_topic | False |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Protein Binding |
| mesh[14].qualifier_ui | Q000378 |
| mesh[14].descriptor_ui | D008099 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | metabolism |
| mesh[14].descriptor_name | Liver |
| mesh[15].qualifier_ui | Q000378 |
| mesh[15].descriptor_ui | D051557 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | metabolism |
| mesh[15].descriptor_name | Hepatocyte Nuclear Factor 4 |
| mesh[16].qualifier_ui | Q000235 |
| mesh[16].descriptor_ui | D051557 |
| mesh[16].is_major_topic | False |
| mesh[16].qualifier_name | genetics |
| mesh[16].descriptor_name | Hepatocyte Nuclear Factor 4 |
| mesh[17].qualifier_ui | Q000378 |
| mesh[17].descriptor_ui | D022762 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | metabolism |
| mesh[17].descriptor_name | CCAAT-Enhancer-Binding Proteins |
| mesh[18].qualifier_ui | Q000235 |
| mesh[18].descriptor_ui | D022762 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | genetics |
| mesh[18].descriptor_name | CCAAT-Enhancer-Binding Proteins |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D000818 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Animals |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D019175 |
| mesh[20].is_major_topic | True |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | DNA Methylation |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D001665 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Binding Sites |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D006801 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Humans |
| mesh[23].qualifier_ui | Q000378 |
| mesh[23].descriptor_ui | D014157 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | metabolism |
| mesh[23].descriptor_name | Transcription Factors |
| mesh[24].qualifier_ui | Q000235 |
| mesh[24].descriptor_ui | D014157 |
| mesh[24].is_major_topic | True |
| mesh[24].qualifier_name | genetics |
| mesh[24].descriptor_name | Transcription Factors |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D019143 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Evolution, Molecular |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D051379 |
| mesh[26].is_major_topic | False |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | Mice |
| mesh[27].qualifier_ui | |
| mesh[27].descriptor_ui | D051381 |
| mesh[27].is_major_topic | False |
| mesh[27].qualifier_name | |
| mesh[27].descriptor_name | Rats |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D018899 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | CpG Islands |
| mesh[29].qualifier_ui | |
| mesh[29].descriptor_ui | D004285 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | |
| mesh[29].descriptor_name | Dogs |
| mesh[30].qualifier_ui | Q000378 |
| mesh[30].descriptor_ui | D051541 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | metabolism |
| mesh[30].descriptor_name | Hepatocyte Nuclear Factor 3-alpha |
| mesh[31].qualifier_ui | Q000235 |
| mesh[31].descriptor_ui | D051541 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | genetics |
| mesh[31].descriptor_name | Hepatocyte Nuclear Factor 3-alpha |
| mesh[32].qualifier_ui | |
| mesh[32].descriptor_ui | D011485 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | |
| mesh[32].descriptor_name | Protein Binding |
| mesh[33].qualifier_ui | Q000378 |
| mesh[33].descriptor_ui | D008099 |
| mesh[33].is_major_topic | False |
| mesh[33].qualifier_name | metabolism |
| mesh[33].descriptor_name | Liver |
| mesh[34].qualifier_ui | Q000378 |
| mesh[34].descriptor_ui | D051557 |
| mesh[34].is_major_topic | False |
| mesh[34].qualifier_name | metabolism |
| mesh[34].descriptor_name | Hepatocyte Nuclear Factor 4 |
| mesh[35].qualifier_ui | Q000235 |
| mesh[35].descriptor_ui | D051557 |
| mesh[35].is_major_topic | False |
| mesh[35].qualifier_name | genetics |
| mesh[35].descriptor_name | Hepatocyte Nuclear Factor 4 |
| mesh[36].qualifier_ui | Q000378 |
| mesh[36].descriptor_ui | D022762 |
| mesh[36].is_major_topic | False |
| mesh[36].qualifier_name | metabolism |
| mesh[36].descriptor_name | CCAAT-Enhancer-Binding Proteins |
| mesh[37].qualifier_ui | Q000235 |
| mesh[37].descriptor_ui | D022762 |
| mesh[37].is_major_topic | False |
| mesh[37].qualifier_name | genetics |
| mesh[37].descriptor_name | CCAAT-Enhancer-Binding Proteins |
| mesh[38].qualifier_ui | |
| mesh[38].descriptor_ui | D000818 |
| mesh[38].is_major_topic | False |
| mesh[38].qualifier_name | |
| mesh[38].descriptor_name | Animals |
| mesh[39].qualifier_ui | |
| mesh[39].descriptor_ui | D019175 |
| mesh[39].is_major_topic | True |
| mesh[39].qualifier_name | |
| mesh[39].descriptor_name | DNA Methylation |
| mesh[40].qualifier_ui | |
| mesh[40].descriptor_ui | D001665 |
| mesh[40].is_major_topic | False |
| mesh[40].qualifier_name | |
| mesh[40].descriptor_name | Binding Sites |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D006801 |
| mesh[41].is_major_topic | False |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | Humans |
| mesh[42].qualifier_ui | Q000378 |
| mesh[42].descriptor_ui | D014157 |
| mesh[42].is_major_topic | True |
| mesh[42].qualifier_name | metabolism |
| mesh[42].descriptor_name | Transcription Factors |
| mesh[43].qualifier_ui | Q000235 |
| mesh[43].descriptor_ui | D014157 |
| mesh[43].is_major_topic | True |
| mesh[43].qualifier_name | genetics |
| mesh[43].descriptor_name | Transcription Factors |
| mesh[44].qualifier_ui | |
| mesh[44].descriptor_ui | D019143 |
| mesh[44].is_major_topic | True |
| mesh[44].qualifier_name | |
| mesh[44].descriptor_name | Evolution, Molecular |
| mesh[45].qualifier_ui | |
| mesh[45].descriptor_ui | D051379 |
| mesh[45].is_major_topic | False |
| mesh[45].qualifier_name | |
| mesh[45].descriptor_name | Mice |
| mesh[46].qualifier_ui | |
| mesh[46].descriptor_ui | D051381 |
| mesh[46].is_major_topic | False |
| mesh[46].qualifier_name | |
| mesh[46].descriptor_name | Rats |
| mesh[47].qualifier_ui | |
| mesh[47].descriptor_ui | D018899 |
| mesh[47].is_major_topic | False |
| mesh[47].qualifier_name | |
| mesh[47].descriptor_name | CpG Islands |
| mesh[48].qualifier_ui | |
| mesh[48].descriptor_ui | D004285 |
| mesh[48].is_major_topic | False |
| mesh[48].qualifier_name | |
| mesh[48].descriptor_name | Dogs |
| mesh[49].qualifier_ui | Q000378 |
| mesh[49].descriptor_ui | D051541 |
| mesh[49].is_major_topic | False |
| mesh[49].qualifier_name | metabolism |
| mesh[49].descriptor_name | Hepatocyte Nuclear Factor 3-alpha |
| type | article |
| title | DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contexts |
| awards[0].id | https://openalex.org/G4529206470 |
| awards[0].funder_id | https://openalex.org/F4320311904 |
| awards[0].funder_award_id | WT202878/Z/16/Z |
| awards[0].funder_display_name | Wellcome Trust |
| awards[1].id | https://openalex.org/G4761255497 |
| awards[1].funder_id | https://openalex.org/F4320311904 |
| awards[1].funder_award_id | WT202878/B/16/Z |
| awards[1].funder_display_name | Wellcome Trust |
| awards[2].id | https://openalex.org/G7143909994 |
| awards[2].funder_id | https://openalex.org/F4320338335 |
| awards[2].funder_award_id | 615584 |
| awards[2].funder_display_name | H2020 European Research Council |
| awards[3].id | https://openalex.org/G376393528 |
| awards[3].funder_id | https://openalex.org/F4320338335 |
| awards[3].funder_award_id | 788937 |
| awards[3].funder_display_name | H2020 European Research Council |
| awards[4].id | https://openalex.org/G776593930 |
| awards[4].funder_id | https://openalex.org/F4320311904 |
| awards[4].funder_award_id | WT108749/Z/15/Z |
| awards[4].funder_display_name | Wellcome Trust |
| biblio.issue | 1 |
| biblio.volume | 25 |
| biblio.last_page | 146 |
| biblio.first_page | 146 |
| grants[0].funder | https://openalex.org/F4320307844 |
| grants[0].award_id | |
| grants[0].funder_display_name | European Molecular Biology Organization |
| grants[1].funder | https://openalex.org/F4320311904 |
| grants[1].award_id | WT108749/Z/15/Z |
| grants[1].funder_display_name | Wellcome Trust |
| grants[2].funder | https://openalex.org/F4320311904 |
| grants[2].award_id | WT202878/B/16/Z |
| grants[2].funder_display_name | Wellcome Trust |
| grants[3].funder | https://openalex.org/F4320311904 |
| grants[3].award_id | WT202878/Z/16/Z |
| grants[3].funder_display_name | Wellcome Trust |
| grants[4].funder | https://openalex.org/F4320338335 |
| grants[4].award_id | 615584 |
| grants[4].funder_display_name | H2020 European Research Council |
| grants[5].funder | https://openalex.org/F4320338335 |
| grants[5].award_id | 788937 |
| grants[5].funder_display_name | H2020 European Research Council |
| topics[0].id | https://openalex.org/T10269 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Epigenetics and DNA Methylation |
| topics[1].id | https://openalex.org/T10222 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9646999835968018 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Genomics and Chromatin Dynamics |
| topics[2].id | https://openalex.org/T11928 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9506000280380249 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genetic Syndromes and Imprinting |
| funders[0].id | https://openalex.org/F4320307844 |
| funders[0].ror | https://ror.org/04wfr2810 |
| funders[0].display_name | European Molecular Biology Organization |
| funders[1].id | https://openalex.org/F4320311904 |
| funders[1].ror | https://ror.org/029chgv08 |
| funders[1].display_name | Wellcome Trust |
| funders[2].id | https://openalex.org/F4320338335 |
| funders[2].ror | https://ror.org/0472cxd90 |
| funders[2].display_name | H2020 European Research Council |
| is_xpac | False |
| apc_list.value | 4090 |
| apc_list.currency | EUR |
| apc_list.value_usd | 5030 |
| apc_paid.value | 4090 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 5030 |
| concepts[0].id | https://openalex.org/C190727270 |
| concepts[0].level | 4 |
| concepts[0].score | 0.8212659358978271 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q874745 |
| concepts[0].display_name | DNA methylation |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.8093488216400146 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C3662595 |
| concepts[2].level | 5 |
| concepts[2].score | 0.7115248441696167 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q5205743 |
| concepts[2].display_name | DNA binding site |
| concepts[3].id | https://openalex.org/C121912465 |
| concepts[3].level | 5 |
| concepts[3].score | 0.6730403900146484 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q3589153 |
| concepts[3].display_name | Epigenomics |
| concepts[4].id | https://openalex.org/C54355233 |
| concepts[4].level | 1 |
| concepts[4].score | 0.6414521932601929 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[4].display_name | Genetics |
| concepts[5].id | https://openalex.org/C33288867 |
| concepts[5].level | 3 |
| concepts[5].score | 0.625361442565918 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q518328 |
| concepts[5].display_name | Methylation |
| concepts[6].id | https://openalex.org/C41091548 |
| concepts[6].level | 3 |
| concepts[6].score | 0.5642117261886597 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q26939 |
| concepts[6].display_name | Epigenetics |
| concepts[7].id | https://openalex.org/C163002167 |
| concepts[7].level | 5 |
| concepts[7].score | 0.4921342730522156 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7277124 |
| concepts[7].display_name | RNA-Directed DNA Methylation |
| concepts[8].id | https://openalex.org/C2778760011 |
| concepts[8].level | 5 |
| concepts[8].score | 0.47453439235687256 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q21163346 |
| concepts[8].display_name | CTCF |
| concepts[9].id | https://openalex.org/C86339819 |
| concepts[9].level | 3 |
| concepts[9].score | 0.4700174629688263 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q407384 |
| concepts[9].display_name | Transcription factor |
| concepts[10].id | https://openalex.org/C83640560 |
| concepts[10].level | 3 |
| concepts[10].score | 0.42966002225875854 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q180951 |
| concepts[10].display_name | Chromatin |
| concepts[11].id | https://openalex.org/C552990157 |
| concepts[11].level | 2 |
| concepts[11].score | 0.34939810633659363 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7430 |
| concepts[11].display_name | DNA |
| concepts[12].id | https://openalex.org/C70721500 |
| concepts[12].level | 1 |
| concepts[12].score | 0.3410235643386841 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[12].display_name | Computational biology |
| concepts[13].id | https://openalex.org/C104317684 |
| concepts[13].level | 2 |
| concepts[13].score | 0.20827704668045044 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[13].display_name | Gene |
| concepts[14].id | https://openalex.org/C101762097 |
| concepts[14].level | 4 |
| concepts[14].score | 0.20275133848190308 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q224093 |
| concepts[14].display_name | Promoter |
| concepts[15].id | https://openalex.org/C111936080 |
| concepts[15].level | 4 |
| concepts[15].score | 0.09081575274467468 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q913367 |
| concepts[15].display_name | Enhancer |
| concepts[16].id | https://openalex.org/C150194340 |
| concepts[16].level | 3 |
| concepts[16].score | 0.08475974202156067 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[16].display_name | Gene expression |
| keywords[0].id | https://openalex.org/keywords/dna-methylation |
| keywords[0].score | 0.8212659358978271 |
| keywords[0].display_name | DNA methylation |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.8093488216400146 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/dna-binding-site |
| keywords[2].score | 0.7115248441696167 |
| keywords[2].display_name | DNA binding site |
| keywords[3].id | https://openalex.org/keywords/epigenomics |
| keywords[3].score | 0.6730403900146484 |
| keywords[3].display_name | Epigenomics |
| keywords[4].id | https://openalex.org/keywords/genetics |
| keywords[4].score | 0.6414521932601929 |
| keywords[4].display_name | Genetics |
| keywords[5].id | https://openalex.org/keywords/methylation |
| keywords[5].score | 0.625361442565918 |
| keywords[5].display_name | Methylation |
| keywords[6].id | https://openalex.org/keywords/epigenetics |
| keywords[6].score | 0.5642117261886597 |
| keywords[6].display_name | Epigenetics |
| keywords[7].id | https://openalex.org/keywords/rna-directed-dna-methylation |
| keywords[7].score | 0.4921342730522156 |
| keywords[7].display_name | RNA-Directed DNA Methylation |
| keywords[8].id | https://openalex.org/keywords/ctcf |
| keywords[8].score | 0.47453439235687256 |
| keywords[8].display_name | CTCF |
| keywords[9].id | https://openalex.org/keywords/transcription-factor |
| keywords[9].score | 0.4700174629688263 |
| keywords[9].display_name | Transcription factor |
| keywords[10].id | https://openalex.org/keywords/chromatin |
| keywords[10].score | 0.42966002225875854 |
| keywords[10].display_name | Chromatin |
| keywords[11].id | https://openalex.org/keywords/dna |
| keywords[11].score | 0.34939810633659363 |
| keywords[11].display_name | DNA |
| keywords[12].id | https://openalex.org/keywords/computational-biology |
| keywords[12].score | 0.3410235643386841 |
| keywords[12].display_name | Computational biology |
| keywords[13].id | https://openalex.org/keywords/gene |
| keywords[13].score | 0.20827704668045044 |
| keywords[13].display_name | Gene |
| keywords[14].id | https://openalex.org/keywords/promoter |
| keywords[14].score | 0.20275133848190308 |
| keywords[14].display_name | Promoter |
| keywords[15].id | https://openalex.org/keywords/enhancer |
| keywords[15].score | 0.09081575274467468 |
| keywords[15].display_name | Enhancer |
| keywords[16].id | https://openalex.org/keywords/gene-expression |
| keywords[16].score | 0.08475974202156067 |
| keywords[16].display_name | Gene expression |
| language | en |
| locations[0].id | doi:10.1186/s13059-024-03218-6 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S81160022 |
| locations[0].source.issn | 1474-7596, 1474-760X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1474-7596 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Genome biology |
| locations[0].source.host_organization | https://openalex.org/P4310320256 |
| locations[0].source.host_organization_name | BioMed Central |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320256 |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6 |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Genome Biology |
| locations[0].landing_page_url | https://doi.org/10.1186/s13059-024-03218-6 |
| locations[1].id | pmid:38844976 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Genome biology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/38844976 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:11155190 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | publishedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | True |
| locations[2].is_published | True |
| locations[2].raw_source_name | Genome Biol |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11155190 |
| locations[3].id | pmh:oai:qmro.qmul.ac.uk:123456789/97366 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306400530 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Queen Mary Research Online (Queen Mary University of London) |
| locations[3].source.host_organization | https://openalex.org/I166337079 |
| locations[3].source.host_organization_name | Queen Mary University of London |
| locations[3].source.host_organization_lineage | https://openalex.org/I166337079 |
| locations[3].license | cc-by |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Article |
| locations[3].license_id | https://openalex.org/licenses/cc-by |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | https://qmro.qmul.ac.uk/xmlui/handle/123456789/97366 |
| locations[4].id | pmh:oai:doaj.org/article:cf477576d182432584630d6c8a3a968c |
| locations[4].is_oa | False |
| locations[4].source.id | https://openalex.org/S4306401280 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[4].source.host_organization | |
| locations[4].source.host_organization_name | |
| locations[4].source.host_organization_lineage | |
| locations[4].license | |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | article |
| locations[4].license_id | |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Genome Biology, Vol 25, Iss 1, Pp 1-22 (2024) |
| locations[4].landing_page_url | https://doaj.org/article/cf477576d182432584630d6c8a3a968c |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5058731375 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-5041-3194 |
| authorships[0].author.display_name | Martina Rimoldi |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I1303153112 |
| authorships[0].affiliations[0].raw_affiliation_string | European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK |
| authorships[0].institutions[0].id | https://openalex.org/I1303153112 |
| authorships[0].institutions[0].ror | https://ror.org/02catss52 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I1303153112, https://openalex.org/I4210138560 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | European Bioinformatics Institute |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Martina Rimoldi |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK |
| authorships[1].author.id | https://openalex.org/A5103480818 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Ning Wang |
| authorships[1].countries | SE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Medical Biochemistry and Biophysics, Division of Functional Genomics and Systems Biology, Karolinska Institutet, Stockholm, SE, 141 83, Sweden |
| authorships[1].institutions[0].id | https://openalex.org/I28166907 |
| authorships[1].institutions[0].ror | https://ror.org/056d84691 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[1].institutions[0].country_code | SE |
| authorships[1].institutions[0].display_name | Karolinska Institutet |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Ning Wang |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Medical Biochemistry and Biophysics, Division of Functional Genomics and Systems Biology, Karolinska Institutet, Stockholm, SE, 141 83, Sweden |
| authorships[2].author.id | https://openalex.org/A5100624322 |
| authorships[2].author.orcid | https://orcid.org/0000-0002-9976-1605 |
| authorships[2].author.display_name | Jilin Zhang |
| authorships[2].countries | SE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Medical Biochemistry and Biophysics, Division of Functional Genomics and Systems Biology, Karolinska Institutet, Stockholm, SE, 141 83, Sweden |
| authorships[2].institutions[0].id | https://openalex.org/I28166907 |
| authorships[2].institutions[0].ror | https://ror.org/056d84691 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[2].institutions[0].country_code | SE |
| authorships[2].institutions[0].display_name | Karolinska Institutet |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Jilin Zhang |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Medical Biochemistry and Biophysics, Division of Functional Genomics and Systems Biology, Karolinska Institutet, Stockholm, SE, 141 83, Sweden |
| authorships[3].author.id | https://openalex.org/A5084501186 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-8614-6232 |
| authorships[3].author.display_name | Diego Villar |
| authorships[3].countries | GB |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I241749, https://openalex.org/I2801316944 |
| authorships[3].affiliations[0].raw_affiliation_string | Cancer Research UK Cambridge Institute, University of Cambridge, Robinson Way, Cambridge, 0RE, CB2, UK |
| authorships[3].institutions[0].id | https://openalex.org/I2801316944 |
| authorships[3].institutions[0].ror | https://ror.org/054225q67 |
| authorships[3].institutions[0].type | nonprofit |
| authorships[3].institutions[0].lineage | https://openalex.org/I2801316944 |
| authorships[3].institutions[0].country_code | GB |
| authorships[3].institutions[0].display_name | Cancer Research UK |
| authorships[3].institutions[1].id | https://openalex.org/I241749 |
| authorships[3].institutions[1].ror | https://ror.org/013meh722 |
| authorships[3].institutions[1].type | education |
| authorships[3].institutions[1].lineage | https://openalex.org/I241749 |
| authorships[3].institutions[1].country_code | GB |
| authorships[3].institutions[1].display_name | University of Cambridge |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Diego Villar |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Cancer Research UK Cambridge Institute, University of Cambridge, Robinson Way, Cambridge, 0RE, CB2, UK |
| authorships[4].author.id | https://openalex.org/A5056137343 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-6201-5599 |
| authorships[4].author.display_name | Duncan T. Odom |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I241749, https://openalex.org/I2801316944 |
| authorships[4].affiliations[0].raw_affiliation_string | Cancer Research UK Cambridge Institute, University of Cambridge, Robinson Way, Cambridge, 0RE, CB2, UK |
| authorships[4].institutions[0].id | https://openalex.org/I2801316944 |
| authorships[4].institutions[0].ror | https://ror.org/054225q67 |
| authorships[4].institutions[0].type | nonprofit |
| authorships[4].institutions[0].lineage | https://openalex.org/I2801316944 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | Cancer Research UK |
| authorships[4].institutions[1].id | https://openalex.org/I241749 |
| authorships[4].institutions[1].ror | https://ror.org/013meh722 |
| authorships[4].institutions[1].type | education |
| authorships[4].institutions[1].lineage | https://openalex.org/I241749 |
| authorships[4].institutions[1].country_code | GB |
| authorships[4].institutions[1].display_name | University of Cambridge |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Duncan T. Odom |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Cancer Research UK Cambridge Institute, University of Cambridge, Robinson Way, Cambridge, 0RE, CB2, UK |
| authorships[5].author.id | https://openalex.org/A5031611528 |
| authorships[5].author.orcid | https://orcid.org/0000-0003-4204-0951 |
| authorships[5].author.display_name | Jussi Taipale |
| authorships[5].countries | SE |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I28166907 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Medical Biochemistry and Biophysics, Division of Functional Genomics and Systems Biology, Karolinska Institutet, Stockholm, SE, 141 83, Sweden |
| authorships[5].institutions[0].id | https://openalex.org/I28166907 |
| authorships[5].institutions[0].ror | https://ror.org/056d84691 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I28166907 |
| authorships[5].institutions[0].country_code | SE |
| authorships[5].institutions[0].display_name | Karolinska Institutet |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Jussi Taipale |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Medical Biochemistry and Biophysics, Division of Functional Genomics and Systems Biology, Karolinska Institutet, Stockholm, SE, 141 83, Sweden |
| authorships[6].author.id | https://openalex.org/A5006816272 |
| authorships[6].author.orcid | https://orcid.org/0000-0002-3897-7955 |
| authorships[6].author.display_name | Paul Flicek |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1303153112 |
| authorships[6].affiliations[0].raw_affiliation_string | European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK |
| authorships[6].institutions[0].id | https://openalex.org/I1303153112 |
| authorships[6].institutions[0].ror | https://ror.org/02catss52 |
| authorships[6].institutions[0].type | facility |
| authorships[6].institutions[0].lineage | https://openalex.org/I1303153112, https://openalex.org/I4210138560 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | European Bioinformatics Institute |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Paul Flicek |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK |
| authorships[7].author.id | https://openalex.org/A5048615517 |
| authorships[7].author.orcid | https://orcid.org/0000-0001-5667-3317 |
| authorships[7].author.display_name | Maša Roller |
| authorships[7].countries | GB |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I1303153112 |
| authorships[7].affiliations[0].raw_affiliation_string | European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK |
| authorships[7].institutions[0].id | https://openalex.org/I1303153112 |
| authorships[7].institutions[0].ror | https://ror.org/02catss52 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I1303153112, https://openalex.org/I4210138560 |
| authorships[7].institutions[0].country_code | GB |
| authorships[7].institutions[0].display_name | European Bioinformatics Institute |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Maša Roller |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contexts |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10269 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Epigenetics and DNA Methylation |
| related_works | https://openalex.org/W2000109482, https://openalex.org/W2019343612, https://openalex.org/W2100995605, https://openalex.org/W2005026459, https://openalex.org/W2162436860, https://openalex.org/W4296785489, https://openalex.org/W4388841077, https://openalex.org/W2114970231, https://openalex.org/W4229066863, https://openalex.org/W2150628237 |
| cited_by_count | 21 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 15 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 3 |
| counts_by_year[2].year | 2023 |
| counts_by_year[2].cited_by_count | 3 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1186/s13059-024-03218-6 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S81160022 |
| best_oa_location.source.issn | 1474-7596, 1474-760X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1474-7596 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Genome biology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320256 |
| best_oa_location.source.host_organization_name | BioMed Central |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320256 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6 |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Genome Biology |
| best_oa_location.landing_page_url | https://doi.org/10.1186/s13059-024-03218-6 |
| primary_location.id | doi:10.1186/s13059-024-03218-6 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S81160022 |
| primary_location.source.issn | 1474-7596, 1474-760X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1474-7596 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Genome biology |
| primary_location.source.host_organization | https://openalex.org/P4310320256 |
| primary_location.source.host_organization_name | BioMed Central |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320256 |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://genomebiology.biomedcentral.com/counter/pdf/10.1186/s13059-024-03218-6 |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Genome Biology |
| primary_location.landing_page_url | https://doi.org/10.1186/s13059-024-03218-6 |
| publication_date | 2024-06-06 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W2790116454, https://openalex.org/W2038584294, https://openalex.org/W1991543373, https://openalex.org/W2160953907, https://openalex.org/W1988871238, https://openalex.org/W2901901698, https://openalex.org/W2139367243, https://openalex.org/W2752520383, https://openalex.org/W2950454586, https://openalex.org/W2095005830, https://openalex.org/W2161350677, https://openalex.org/W2065103652, https://openalex.org/W2954492537, https://openalex.org/W1983275320, https://openalex.org/W2481378334, https://openalex.org/W2950817352, https://openalex.org/W3009045514, https://openalex.org/W2995813047, https://openalex.org/W2084203180, https://openalex.org/W2039053594, https://openalex.org/W2581467944, https://openalex.org/W2205615102, https://openalex.org/W2976009347, https://openalex.org/W2953367677, https://openalex.org/W2979330888, https://openalex.org/W2968658941, https://openalex.org/W2123032740, https://openalex.org/W2114970231, https://openalex.org/W2012462525, https://openalex.org/W2031562994, https://openalex.org/W2141816956, https://openalex.org/W2327450572, https://openalex.org/W2942230700, https://openalex.org/W2494121918, https://openalex.org/W2610903912, https://openalex.org/W2625488274, https://openalex.org/W2128059998, https://openalex.org/W1978897662, https://openalex.org/W2119347606, https://openalex.org/W1506343731, https://openalex.org/W2171808845, https://openalex.org/W2166159761, https://openalex.org/W2086038224, https://openalex.org/W2982555675, https://openalex.org/W2792883187, https://openalex.org/W2588517583, https://openalex.org/W3130610799, https://openalex.org/W2281531600, https://openalex.org/W2072063319, https://openalex.org/W2219596071, https://openalex.org/W1981354816, https://openalex.org/W2336968766, https://openalex.org/W2952170484, https://openalex.org/W2095903180, https://openalex.org/W2810869921, https://openalex.org/W1699926620, https://openalex.org/W4205247356, https://openalex.org/W2131374955, https://openalex.org/W2170551349, https://openalex.org/W2102619694, https://openalex.org/W1709815505, https://openalex.org/W4242289937, https://openalex.org/W2938574745, https://openalex.org/W3016522754, https://openalex.org/W3031159512, https://openalex.org/W2983170455 |
| referenced_works_count | 66 |
| abstract_inverted_index.a | 60, 151, 230 |
| abstract_inverted_index.TF | 48, 71, 110, 167, 207, 236, 248 |
| abstract_inverted_index.an | 5 |
| abstract_inverted_index.at | 166 |
| abstract_inverted_index.by | 129 |
| abstract_inverted_index.in | 13, 27, 32, 74, 98, 135, 235 |
| abstract_inverted_index.is | 4 |
| abstract_inverted_index.of | 67, 89, 101, 133, 162, 191, 206 |
| abstract_inverted_index.to | 63, 70 |
| abstract_inverted_index.we | 58, 77, 156, 198 |
| abstract_inverted_index.DNA | 2, 68, 80, 163, 200, 233, 244 |
| abstract_inverted_index.Its | 17 |
| abstract_inverted_index.Our | 226 |
| abstract_inverted_index.TFs | 92 |
| abstract_inverted_index.and | 38, 47, 82, 153, 159, 173, 188, 250, 259 |
| abstract_inverted_index.are | 19, 53, 113, 127, 143, 251 |
| abstract_inverted_index.but | 30, 116 |
| abstract_inverted_index.for | 232 |
| abstract_inverted_index.has | 10, 216 |
| abstract_inverted_index.its | 217, 222 |
| abstract_inverted_index.low | 31 |
| abstract_inverted_index.our | 195 |
| abstract_inverted_index.own | 218 |
| abstract_inverted_index.the | 23, 43, 65, 99, 130, 146, 176 |
| abstract_inverted_index.use | 59 |
| abstract_inverted_index.(TF) | 35 |
| abstract_inverted_index.CpGs | 134, 142 |
| abstract_inverted_index.Here | 57 |
| abstract_inverted_index.also | 119 |
| abstract_inverted_index.core | 147 |
| abstract_inverted_index.each | 213 |
| abstract_inverted_index.even | 140 |
| abstract_inverted_index.find | 199 |
| abstract_inverted_index.five | 90, 102 |
| abstract_inverted_index.from | 145 |
| abstract_inverted_index.gain | 202 |
| abstract_inverted_index.have | 120 |
| abstract_inverted_index.high | 26 |
| abstract_inverted_index.loss | 205 |
| abstract_inverted_index.rat, | 108 |
| abstract_inverted_index.role | 231 |
| abstract_inverted_index.same | 177 |
| abstract_inverted_index.site | 50 |
| abstract_inverted_index.that | 242 |
| abstract_inverted_index.they | 117 |
| abstract_inverted_index.this | 84 |
| abstract_inverted_index.upon | 203 |
| abstract_inverted_index.when | 141 |
| abstract_inverted_index.with | 85, 253 |
| abstract_inverted_index.FOXA1 | 174 |
| abstract_inverted_index.These | 183 |
| abstract_inverted_index.dog). | 109 |
| abstract_inverted_index.liver | 100 |
| abstract_inverted_index.lowly | 114 |
| abstract_inverted_index.often | 118 |
| abstract_inverted_index.roles | 12 |
| abstract_inverted_index.share | 175 |
| abstract_inverted_index.sites | 37, 112, 126 |
| abstract_inverted_index.still | 54 |
| abstract_inverted_index.their | 136, 254 |
| abstract_inverted_index.which | 9 |
| abstract_inverted_index.while | 180 |
| abstract_inverted_index.wider | 137 |
| abstract_inverted_index.(CTCF, | 93 |
| abstract_inverted_index.CEBPA, | 94, 170 |
| abstract_inverted_index.CTCF's | 181 |
| abstract_inverted_index.FOXA1) | 97 |
| abstract_inverted_index.HNF4A, | 95, 171 |
| abstract_inverted_index.absent | 144 |
| abstract_inverted_index.across | 22, 239 |
| abstract_inverted_index.active | 39 |
| abstract_inverted_index.biding | 125 |
| abstract_inverted_index.factor | 34 |
| abstract_inverted_index.genome | 15 |
| abstract_inverted_index.levels | 18, 165 |
| abstract_inverted_index.motif. | 149 |
| abstract_inverted_index.mouse, | 107 |
| abstract_inverted_index.status | 132 |
| abstract_inverted_index.(human, | 105 |
| abstract_inverted_index.Results | 56 |
| abstract_inverted_index.binding | 36, 49, 72, 111, 138, 148, 168, 237, 249 |
| abstract_inverted_index.changes | 238 |
| abstract_inverted_index.combine | 83 |
| abstract_inverted_index.differ. | 182 |
| abstract_inverted_index.extract | 157 |
| abstract_inverted_index.genome, | 24 |
| abstract_inverted_index.genomic | 223 |
| abstract_inverted_index.levels. | 123 |
| abstract_inverted_index.pattern | 215 |
| abstract_inverted_index.profile | 79 |
| abstract_inverted_index.regions | 29, 139 |
| abstract_inverted_index.species | 104, 240 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.However, | 42 |
| abstract_inverted_index.ONECUT1, | 96, 172 |
| abstract_inverted_index.TF-bound | 192 |
| abstract_inverted_index.analyses | 228 |
| abstract_inverted_index.approach | 62 |
| abstract_inverted_index.distinct | 91, 158 |
| abstract_inverted_index.indicate | 229 |
| abstract_inverted_index.macaque, | 106 |
| abstract_inverted_index.mammals. | 75 |
| abstract_inverted_index.numerous | 11 |
| abstract_inverted_index.patterns | 52, 161, 184 |
| abstract_inverted_index.profiles | 88, 246 |
| abstract_inverted_index.regions. | 41, 169, 193 |
| abstract_inverted_index.specific | 243 |
| abstract_inverted_index.unclear. | 55 |
| abstract_inverted_index.Employing | 150 |
| abstract_inverted_index.activity, | 256 |
| abstract_inverted_index.approach, | 155 |
| abstract_inverted_index.chromatin | 189, 257 |
| abstract_inverted_index.contexts, | 258 |
| abstract_inverted_index.contexts. | 224 |
| abstract_inverted_index.evolution | 73 |
| abstract_inverted_index.function. | 16 |
| abstract_inverted_index.functions | 187 |
| abstract_inverted_index.important | 6 |
| abstract_inverted_index.including | 241 |
| abstract_inverted_index.mammalian | 103 |
| abstract_inverted_index.occupancy | 87 |
| abstract_inverted_index.patterns, | 179 |
| abstract_inverted_index.published | 86 |
| abstract_inverted_index.repressed | 28 |
| abstract_inverted_index.spatially | 20 |
| abstract_inverted_index.typically | 25 |
| abstract_inverted_index.Background | 1 |
| abstract_inverted_index.Leveraging | 194 |
| abstract_inverted_index.associated | 252 |
| abstract_inverted_index.clustering | 154 |
| abstract_inverted_index.correlated | 21 |
| abstract_inverted_index.epigenetic | 7 |
| abstract_inverted_index.epigenomic | 227 |
| abstract_inverted_index.evolution. | 211 |
| abstract_inverted_index.framework, | 197 |
| abstract_inverted_index.indicating | 209 |
| abstract_inverted_index.influenced | 128 |
| abstract_inverted_index.landscapes | 190 |
| abstract_inverted_index.mechanisms | 44 |
| abstract_inverted_index.modulating | 14 |
| abstract_inverted_index.occupancy, | 208 |
| abstract_inverted_index.reflecting | 221 |
| abstract_inverted_index.regulatory | 40, 255 |
| abstract_inverted_index.trajectory | 220 |
| abstract_inverted_index.Conclusions | 225 |
| abstract_inverted_index.alternative | 186 |
| abstract_inverted_index.association | 66 |
| abstract_inverted_index.comparative | 61 |
| abstract_inverted_index.coordinated | 210 |
| abstract_inverted_index.genome-wide | 46 |
| abstract_inverted_index.investigate | 64 |
| abstract_inverted_index.methylated, | 115 |
| abstract_inverted_index.methylation | 3, 51, 69, 81, 122, 131, 164, 178, 201, 214, 234, 245 |
| abstract_inverted_index.Furthermore, | 124, 212 |
| abstract_inverted_index.characterize | 185, 247 |
| abstract_inverted_index.establishing | 45 |
| abstract_inverted_index.evolutionary | 204, 219, 260 |
| abstract_inverted_index.intermediate | 121 |
| abstract_inverted_index.modification | 8 |
| abstract_inverted_index.phylogenetic | 196 |
| abstract_inverted_index.Specifically, | 76 |
| abstract_inverted_index.trajectories. | 261 |
| abstract_inverted_index.transcription | 33 |
| abstract_inverted_index.classification | 152 |
| abstract_inverted_index.experimentally | 78 |
| abstract_inverted_index.species-conserved | 160 |
| cited_by_percentile_year.max | 100 |
| cited_by_percentile_year.min | 96 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 8 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.6700000166893005 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.96902359 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |