Dysbiosis Associated With Esophageal Adenocarcinoma—A Novel Method to Detect Tissue-Associated Microbiome Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1016/j.gastha.2022.05.016
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1016/j.gastha.2022.05.016
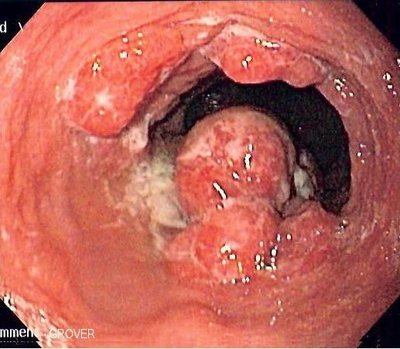
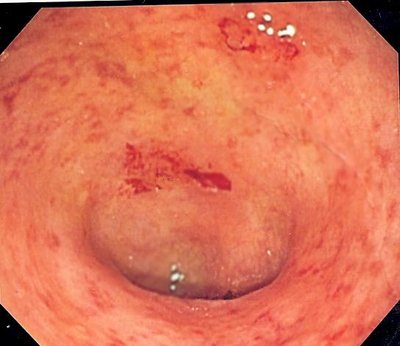
Esophageal cancer (EC) is the eighth most common cancer diagnosed globally and one of the primary drivers of cancer-related deaths.1Ferlay J. Soerjomataram I. Dikshit R. et al.Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012.Int J Cancer. 2015; 136: E359-E386Crossref PubMed Scopus (20333) Google Scholar For the past half-century, the incidence of esophageal adenocarcinoma and its precursor, Barrett's esophagus, has been rapidly growing for reasons that are not properly explained by currently established risk factors. Gastroesophageal reflux disease, obesity, and the microbiome present in the upper gastrointestinal (GI) tract could contribute to the pathogenesis.2Snider E.J. Freedberg D.E. Abrams J.A. Potential role of the microbiome in Barrett's esophagus and esophageal adenocarcinoma.Dig Dis Sci. 2016; 61: 2217-2225Crossref PubMed Scopus (39) Google Scholar A recent study found patients with EC to have dysbiosis in the gut microbiome.3Deng Y. Tang D. Hou P. et al.Dysbiosis of gut microbiota in patients with esophageal cancer.Microb Pathog. 2021; 150: 104709Crossref PubMed Scopus (16) Google Scholar The GI microbiome has been shown to play an essential part in health,4Nejad M.R. Ishaq S. Al Dulaimi D. et al.The role of infectious mediators and gut microbiome in the pathogenesis of celiac disease.Arch Iran Med. 2015; 18: 244-249PubMed Google Scholar, 5Ohkusa T. Okayasu I. Ogihara T. et al.Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis.Gut. 2003; 52: 79-83Crossref PubMed Scopus (189) Google Scholar, 6Kostic A.D. Xavier R.J. Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead.Gastroenterology. 2014; 146: 1489-1499Abstract Full Text Full Text PDF PubMed Scopus (1025) Google Scholar in GI and other intestinal disorders,7Hotamisligil G.S. Inflammation and metabolic disorders.Nature. 2006; 444: 860-867Crossref PubMed Scopus (5975) Google Scholar,8Fujimura K.E. Lynch S.V. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome.Cell Host Microbe. 2015; 17: 592-602Abstract Full Text Full Text PDF PubMed Scopus (238) Google Scholar as well as in several forms of cancer.9Vogtmann E. Goedert J.J. Epidemiologic studies of the human microbiome and cancer.Br J Cancer. 2016; 114: 237-242Crossref PubMed Scopus (109) Google Scholar,10Garrett W.S. Cancer and the microbiota.Science. 2015; 348: 80-86Crossref PubMed Scopus (90) Google Scholar The distal esophagus contains a unique microbiome composed primarily of oral microbiota, which has been found to be altered in Barrett's esophagus and reflux esophagitis,2Snider E.J. Freedberg D.E. Abrams J.A. Potential role of the microbiome in Barrett's esophagus and esophageal adenocarcinoma.Dig Dis Sci. 2016; 61: 2217-2225Crossref PubMed Scopus (39) Google Scholar establishing the association of the microbiome with the pathophysiology of esophageal disorders. The role of the microbiome in esophageal disease progression as related to mucosal dysplasia is not well defined. In this cross-sectional study,11Radani N. Metwaly A. Reitmeier S. et al.Analysis of fecal, salivary and tissue microbiome in Barrett esophagus, dysplasia and esophageal adenocarcinoma.Gastro Hep Adv. 2022; Abstract Full Text Full Text PDF Google Scholar the authors investigated the changes in the upper-GI-tract microbiome in the saliva, esophageal tissue, and feces in healthy patients as well as in patients with dysplasia, Barrett's esophagus, or esophageal adenocarcinoma using a novel method for characterizing the tissue-associated microbiome. They performed simultaneous integrated clinical-pathological and epidemiological correlations. Using the PAXgene fixation technique in conjunction with paraffin embedding of tissue biopsies for microbiome investigation, the authors demonstrated a unique microbial shift in tissue biopsies from esophageal adenocarcinoma patients compared to controls. However, total microbial diversity in salivary and fecal samples did not differ significantly across disease progression. The authors suggested that compared to saliva or the fecal microbiome, tissue-biopsy-linked microbiome has a tight association with esophageal adenocarcinoma. A key finding was the reduction in richness of species in esophageal adenocarcinoma compared to other phenotypes. suggesting a potential role of the mucosa-associated microbiome in the pathogenesis of esophageal adenocarcinoma. However, other studies3Deng Y. Tang D. Hou P. et al.Dysbiosis of gut microbiota in patients with esophageal cancer.Microb Pathog. 2021; 150: 104709Crossref PubMed Scopus (16) Google Scholar published recently have found 39 taxa that are more abundant in the feces of patients, with EC having a greater number of harmful bacteria that may be involved in the progression of carcinoma. Further analysis revealed, among other bacterium, that Lachnospira was found more abundantly in patients with EC and can possibly be used as a microbial biomarker and, possibly, direct treatment for EC. The authors suggested that the mucosa-associated microbiome may interact directly with mucosal cells through surface proteins and metabolites. For example, Fusobacterium nucleatum has been shown to interact directly with mucosal cells in colon cancer to enhance colon cancer. This interaction involves binding to E-cadherin and also activation of toll-like receptor 4 (TLR-4) by lipopolysaccharides. A similar mechanism of TLR-4 activation may be involved in esophageal carcinogenesis. In a rat model of esophageal adenocarcinoma, upregulation of TLR 1–3, 6, 7, and 9 in cancerous tissue was noted compared to normal epithelium. The authors detected abundance of Escherichia coli in tumor tissues but not in the adjacent normal epithelium.12Zaidi A.H. Kelly L.A. Kreft R.E. et al.Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma.BMC Cancer. 2016; 16: 52Crossref PubMed Scopus (54) Google Scholar This study highlights the differences in the mucosa-associated microbiome in esophageal adenocarcinoma. Methods to standardize mucosa-associated microbiome are described in the study and can provide a useful tool for investigators trying to understand the role of the gut microbiome in specific diseases. Limitations of the study include the small number of samples analyzed and a potential confounder utilizing control patients with chronic gastritis. Larger studies are needed to see if assessing the microbiome in this fashion will have a role in clinical decision-making or risk assessment in the diagnosis and management of esophageal adenocarcinoma. Finally, and importantly, it remains to be determined if the dysbiosis is leading to the noted esophageal mucosal changes and subsequent adenocarcinoma or whether the abnormal mucosa favors the colonization of specific microbes. In other words, further study is needed to clarify a causal relationship. Further studies involving large cohorts of patients will need to be done to understand the contribution of the gut microbiome in the pathogenesis of esophageal adenocarcinoma. We acknowledge this article contributes to the excitement in the field. Analysis of Fecal, Salivary, and Tissue Microbiome in Barrett's Esophagus, Dysplasia, and Esophageal AdenocarcinomaGastro Hep AdvancesVol. 1Issue 5PreviewEsophageal adenocarcinoma (EAC) incidence has risen dramatically in the Western countries over the past decades. The underlying reasons are incompletely understood, and shifts in the esophageal microbiome have been postulated to increase predisposition to disease development. Multiple factors including medications, lifestyle, and diet could influence microbiome composition and disease progression. The aim of this study was (1) to identify a feasible method to characterize the tissue-associated microbiome, and (2) to investigate differences in the microbiome of saliva, esophageal tissue, and fecal samples by disease state and validate with 2 external cohorts. Full-Text PDF Open Access
Related Topics
- Type
- editorial
- Language
- en
- Landing Page
- https://doi.org/10.1016/j.gastha.2022.05.016
- OA Status
- gold
- References
- 13
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4281790037
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4281790037Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1016/j.gastha.2022.05.016Digital Object Identifier
- Title
-
Dysbiosis Associated With Esophageal Adenocarcinoma—A Novel Method to Detect Tissue-Associated MicrobiomeWork title
- Type
-
editorialOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-01-01Full publication date if available
- Authors
-
Arun Balasubramaniam, Shanthi SrinivasanList of authors in order
- Landing page
-
https://doi.org/10.1016/j.gastha.2022.05.016Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1016/j.gastha.2022.05.016Direct OA link when available
- Concepts
-
Dysbiosis, Microbiome, Gastroenterology, Barrett's esophagus, Esophageal cancer, Medicine, Disease, Esophagus, Cancer, Ulcerative colitis, Incidence (geometry), Internal medicine, Adenocarcinoma, Bioinformatics, Biology, Physics, OpticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
13Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4281790037 |
|---|---|
| doi | https://doi.org/10.1016/j.gastha.2022.05.016 |
| ids.doi | https://doi.org/10.1016/j.gastha.2022.05.016 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/39131842 |
| ids.openalex | https://openalex.org/W4281790037 |
| fwci | 0.0 |
| type | editorial |
| title | Dysbiosis Associated With Esophageal Adenocarcinoma—A Novel Method to Detect Tissue-Associated Microbiome |
| awards[0].id | https://openalex.org/G7306170249 |
| awards[0].funder_id | https://openalex.org/F4320332161 |
| awards[0].display_name | |
| awards[0].funder_award_id | R01DK080684 |
| awards[0].funder_display_name | National Institutes of Health |
| awards[1].id | https://openalex.org/G104091306 |
| awards[1].funder_id | https://openalex.org/F4320337504 |
| awards[1].display_name | |
| awards[1].funder_award_id | I01BX000136-08 |
| awards[1].funder_display_name | Research and Development |
| awards[2].id | https://openalex.org/G545601102 |
| awards[2].funder_id | https://openalex.org/F4320332161 |
| awards[2].display_name | |
| awards[2].funder_award_id | R01DK044234 |
| awards[2].funder_display_name | National Institutes of Health |
| biblio.issue | 5 |
| biblio.volume | 1 |
| biblio.last_page | 776 |
| biblio.first_page | 775 |
| topics[0].id | https://openalex.org/T10066 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9991000294685364 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Gut microbiota and health |
| topics[1].id | https://openalex.org/T10276 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9941999912261963 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2746 |
| topics[1].subfield.display_name | Surgery |
| topics[1].display_name | Helicobacter pylori-related gastroenterology studies |
| topics[2].id | https://openalex.org/T12528 |
| topics[2].field.id | https://openalex.org/fields/24 |
| topics[2].field.display_name | Immunology and Microbiology |
| topics[2].score | 0.9871000051498413 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2403 |
| topics[2].subfield.display_name | Immunology |
| topics[2].display_name | IL-33, ST2, and ILC Pathways |
| funders[0].id | https://openalex.org/F4320332161 |
| funders[0].ror | https://ror.org/01cwqze88 |
| funders[0].display_name | National Institutes of Health |
| funders[1].id | https://openalex.org/F4320337504 |
| funders[1].ror | https://ror.org/027s68j25 |
| funders[1].display_name | Research and Development |
| is_xpac | False |
| apc_list.value | 2000 |
| apc_list.currency | USD |
| apc_list.value_usd | 2000 |
| apc_paid.value | 2000 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2000 |
| concepts[0].id | https://openalex.org/C2777165150 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7232123017311096 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q269334 |
| concepts[0].display_name | Dysbiosis |
| concepts[1].id | https://openalex.org/C143121216 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6703750491142273 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1330402 |
| concepts[1].display_name | Microbiome |
| concepts[2].id | https://openalex.org/C90924648 |
| concepts[2].level | 1 |
| concepts[2].score | 0.6233770847320557 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q120569 |
| concepts[2].display_name | Gastroenterology |
| concepts[3].id | https://openalex.org/C2778589982 |
| concepts[3].level | 4 |
| concepts[3].score | 0.6159583330154419 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q808892 |
| concepts[3].display_name | Barrett's esophagus |
| concepts[4].id | https://openalex.org/C2779742542 |
| concepts[4].level | 3 |
| concepts[4].score | 0.6038590669631958 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q372701 |
| concepts[4].display_name | Esophageal cancer |
| concepts[5].id | https://openalex.org/C71924100 |
| concepts[5].level | 0 |
| concepts[5].score | 0.5668401718139648 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[5].display_name | Medicine |
| concepts[6].id | https://openalex.org/C2779134260 |
| concepts[6].level | 2 |
| concepts[6].score | 0.541337788105011 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q12136 |
| concepts[6].display_name | Disease |
| concepts[7].id | https://openalex.org/C2777819096 |
| concepts[7].level | 2 |
| concepts[7].score | 0.5401550531387329 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q173710 |
| concepts[7].display_name | Esophagus |
| concepts[8].id | https://openalex.org/C121608353 |
| concepts[8].level | 2 |
| concepts[8].score | 0.5378754138946533 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[8].display_name | Cancer |
| concepts[9].id | https://openalex.org/C2780479503 |
| concepts[9].level | 3 |
| concepts[9].score | 0.5247210264205933 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1477 |
| concepts[9].display_name | Ulcerative colitis |
| concepts[10].id | https://openalex.org/C61511704 |
| concepts[10].level | 2 |
| concepts[10].score | 0.5111272931098938 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q1671857 |
| concepts[10].display_name | Incidence (geometry) |
| concepts[11].id | https://openalex.org/C126322002 |
| concepts[11].level | 1 |
| concepts[11].score | 0.49267321825027466 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[11].display_name | Internal medicine |
| concepts[12].id | https://openalex.org/C2781182431 |
| concepts[12].level | 3 |
| concepts[12].score | 0.49208658933639526 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q356033 |
| concepts[12].display_name | Adenocarcinoma |
| concepts[13].id | https://openalex.org/C60644358 |
| concepts[13].level | 1 |
| concepts[13].score | 0.24022641777992249 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[13].display_name | Bioinformatics |
| concepts[14].id | https://openalex.org/C86803240 |
| concepts[14].level | 0 |
| concepts[14].score | 0.19547736644744873 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[14].display_name | Biology |
| concepts[15].id | https://openalex.org/C121332964 |
| concepts[15].level | 0 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[15].display_name | Physics |
| concepts[16].id | https://openalex.org/C120665830 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q14620 |
| concepts[16].display_name | Optics |
| keywords[0].id | https://openalex.org/keywords/dysbiosis |
| keywords[0].score | 0.7232123017311096 |
| keywords[0].display_name | Dysbiosis |
| keywords[1].id | https://openalex.org/keywords/microbiome |
| keywords[1].score | 0.6703750491142273 |
| keywords[1].display_name | Microbiome |
| keywords[2].id | https://openalex.org/keywords/gastroenterology |
| keywords[2].score | 0.6233770847320557 |
| keywords[2].display_name | Gastroenterology |
| keywords[3].id | https://openalex.org/keywords/barretts-esophagus |
| keywords[3].score | 0.6159583330154419 |
| keywords[3].display_name | Barrett's esophagus |
| keywords[4].id | https://openalex.org/keywords/esophageal-cancer |
| keywords[4].score | 0.6038590669631958 |
| keywords[4].display_name | Esophageal cancer |
| keywords[5].id | https://openalex.org/keywords/medicine |
| keywords[5].score | 0.5668401718139648 |
| keywords[5].display_name | Medicine |
| keywords[6].id | https://openalex.org/keywords/disease |
| keywords[6].score | 0.541337788105011 |
| keywords[6].display_name | Disease |
| keywords[7].id | https://openalex.org/keywords/esophagus |
| keywords[7].score | 0.5401550531387329 |
| keywords[7].display_name | Esophagus |
| keywords[8].id | https://openalex.org/keywords/cancer |
| keywords[8].score | 0.5378754138946533 |
| keywords[8].display_name | Cancer |
| keywords[9].id | https://openalex.org/keywords/ulcerative-colitis |
| keywords[9].score | 0.5247210264205933 |
| keywords[9].display_name | Ulcerative colitis |
| keywords[10].id | https://openalex.org/keywords/incidence |
| keywords[10].score | 0.5111272931098938 |
| keywords[10].display_name | Incidence (geometry) |
| keywords[11].id | https://openalex.org/keywords/internal-medicine |
| keywords[11].score | 0.49267321825027466 |
| keywords[11].display_name | Internal medicine |
| keywords[12].id | https://openalex.org/keywords/adenocarcinoma |
| keywords[12].score | 0.49208658933639526 |
| keywords[12].display_name | Adenocarcinoma |
| keywords[13].id | https://openalex.org/keywords/bioinformatics |
| keywords[13].score | 0.24022641777992249 |
| keywords[13].display_name | Bioinformatics |
| keywords[14].id | https://openalex.org/keywords/biology |
| keywords[14].score | 0.19547736644744873 |
| keywords[14].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1016/j.gastha.2022.05.016 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4220651355 |
| locations[0].source.issn | 2772-5723 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2772-5723 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Gastro Hep Advances |
| locations[0].source.host_organization | https://openalex.org/P4310320990 |
| locations[0].source.host_organization_name | Elsevier BV |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320990 |
| locations[0].source.host_organization_lineage_names | Elsevier BV |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Gastro Hep Advances |
| locations[0].landing_page_url | https://doi.org/10.1016/j.gastha.2022.05.016 |
| locations[1].id | pmid:39131842 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Gastro hep advances |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/39131842 |
| locations[2].id | pmh:oai:doaj.org/article:5f901923e73b454d94b625ee2e0f980f |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Gastro Hep Advances, Vol 1, Iss 5, Pp 775-776 (2022) |
| locations[2].landing_page_url | https://doaj.org/article/5f901923e73b454d94b625ee2e0f980f |
| locations[3].id | pmh:oai:osti.gov:2203152 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306402487 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | OSTI OAI (U.S. Department of Energy Office of Scientific and Technical Information) |
| locations[3].source.host_organization | https://openalex.org/I139351228 |
| locations[3].source.host_organization_name | Office of Scientific and Technical Information |
| locations[3].source.host_organization_lineage | https://openalex.org/I139351228 |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | https://www.osti.gov/biblio/2203152 |
| locations[4].id | pmh:oai:pubmedcentral.nih.gov:11307479 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S2764455111 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | PubMed Central |
| locations[4].source.host_organization | https://openalex.org/I1299303238 |
| locations[4].source.host_organization_name | National Institutes of Health |
| locations[4].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[4].license | other-oa |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/other-oa |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Gastro Hep Adv |
| locations[4].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11307479 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5081164723 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-7802-246X |
| authorships[0].author.display_name | Arun Balasubramaniam |
| authorships[0].countries | US |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210149289 |
| authorships[0].affiliations[0].raw_affiliation_string | Atlanta Veterans Affairs Health Care System, Decatur, Georgia |
| authorships[0].affiliations[1].institution_ids | https://openalex.org/I150468666 |
| authorships[0].affiliations[1].raw_affiliation_string | Division of Digestive Diseases, Emory University School of Medicine, Atlanta, Georgia |
| authorships[0].institutions[0].id | https://openalex.org/I4210149289 |
| authorships[0].institutions[0].ror | https://ror.org/041t78y98 |
| authorships[0].institutions[0].type | healthcare |
| authorships[0].institutions[0].lineage | https://openalex.org/I1322918889, https://openalex.org/I2799886695, https://openalex.org/I4210131075, https://openalex.org/I4210149289 |
| authorships[0].institutions[0].country_code | US |
| authorships[0].institutions[0].display_name | Atlanta VA Health Care System |
| authorships[0].institutions[1].id | https://openalex.org/I150468666 |
| authorships[0].institutions[1].ror | https://ror.org/03czfpz43 |
| authorships[0].institutions[1].type | education |
| authorships[0].institutions[1].lineage | https://openalex.org/I150468666 |
| authorships[0].institutions[1].country_code | US |
| authorships[0].institutions[1].display_name | Emory University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Arun Balasubramaniam |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Atlanta Veterans Affairs Health Care System, Decatur, Georgia, Division of Digestive Diseases, Emory University School of Medicine, Atlanta, Georgia |
| authorships[1].author.id | https://openalex.org/A5091361579 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-6275-2369 |
| authorships[1].author.display_name | Shanthi Srinivasan |
| authorships[1].countries | US |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I150468666 |
| authorships[1].affiliations[0].raw_affiliation_string | Division of Digestive Diseases, Emory University School of Medicine, Atlanta, Georgia |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I4210149289 |
| authorships[1].affiliations[1].raw_affiliation_string | Atlanta Veterans Affairs Health Care System, Decatur, Georgia |
| authorships[1].institutions[0].id | https://openalex.org/I4210149289 |
| authorships[1].institutions[0].ror | https://ror.org/041t78y98 |
| authorships[1].institutions[0].type | healthcare |
| authorships[1].institutions[0].lineage | https://openalex.org/I1322918889, https://openalex.org/I2799886695, https://openalex.org/I4210131075, https://openalex.org/I4210149289 |
| authorships[1].institutions[0].country_code | US |
| authorships[1].institutions[0].display_name | Atlanta VA Health Care System |
| authorships[1].institutions[1].id | https://openalex.org/I150468666 |
| authorships[1].institutions[1].ror | https://ror.org/03czfpz43 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I150468666 |
| authorships[1].institutions[1].country_code | US |
| authorships[1].institutions[1].display_name | Emory University |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | Shanthi Srinivasan |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Atlanta Veterans Affairs Health Care System, Decatur, Georgia, Division of Digestive Diseases, Emory University School of Medicine, Atlanta, Georgia |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1016/j.gastha.2022.05.016 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Dysbiosis Associated With Esophageal Adenocarcinoma—A Novel Method to Detect Tissue-Associated Microbiome |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10066 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9991000294685364 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Gut microbiota and health |
| related_works | https://openalex.org/W2378399001, https://openalex.org/W2381376498, https://openalex.org/W2393318770, https://openalex.org/W2369056725, https://openalex.org/W4385147599, https://openalex.org/W3151390891, https://openalex.org/W3007894746, https://openalex.org/W3214964065, https://openalex.org/W4311366456, https://openalex.org/W4283718758 |
| cited_by_count | 0 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1016/j.gastha.2022.05.016 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4220651355 |
| best_oa_location.source.issn | 2772-5723 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2772-5723 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Gastro Hep Advances |
| best_oa_location.source.host_organization | https://openalex.org/P4310320990 |
| best_oa_location.source.host_organization_name | Elsevier BV |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320990 |
| best_oa_location.source.host_organization_lineage_names | Elsevier BV |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Gastro Hep Advances |
| best_oa_location.landing_page_url | https://doi.org/10.1016/j.gastha.2022.05.016 |
| primary_location.id | doi:10.1016/j.gastha.2022.05.016 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4220651355 |
| primary_location.source.issn | 2772-5723 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2772-5723 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Gastro Hep Advances |
| primary_location.source.host_organization | https://openalex.org/P4310320990 |
| primary_location.source.host_organization_name | Elsevier BV |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320990 |
| primary_location.source.host_organization_lineage_names | Elsevier BV |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Gastro Hep Advances |
| primary_location.landing_page_url | https://doi.org/10.1016/j.gastha.2022.05.016 |
| publication_date | 2022-01-01 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W1757407923, https://openalex.org/W2321331974, https://openalex.org/W3117864613, https://openalex.org/W6675037493, https://openalex.org/W2128343971, https://openalex.org/W2152751713, https://openalex.org/W2000054651, https://openalex.org/W1536527549, https://openalex.org/W2228700263, https://openalex.org/W2073924079, https://openalex.org/W2099478147, https://openalex.org/W4226301715, https://openalex.org/W2260354739 |
| referenced_works_count | 13 |
| abstract_inverted_index.2 | 1127 |
| abstract_inverted_index.4 | 763 |
| abstract_inverted_index.9 | 793 |
| abstract_inverted_index.A | 124, 591, 767 |
| abstract_inverted_index.J | 39, 335 |
| abstract_inverted_index.a | 362, 506, 541, 585, 609, 667, 704, 780, 871, 900, 924, 982, 1098 |
| abstract_inverted_index.39 | 653 |
| abstract_inverted_index.6, | 790 |
| abstract_inverted_index.7, | 791 |
| abstract_inverted_index.A. | 445 |
| abstract_inverted_index.Al | 178 |
| abstract_inverted_index.D. | 140, 180, 240, 627 |
| abstract_inverted_index.E. | 324 |
| abstract_inverted_index.EC | 130, 665, 697 |
| abstract_inverted_index.GI | 163, 267 |
| abstract_inverted_index.I. | 22, 206 |
| abstract_inverted_index.In | 439, 779, 973 |
| abstract_inverted_index.J. | 20 |
| abstract_inverted_index.N. | 443 |
| abstract_inverted_index.P. | 142, 629 |
| abstract_inverted_index.R. | 24 |
| abstract_inverted_index.S. | 177, 447 |
| abstract_inverted_index.T. | 204, 208 |
| abstract_inverted_index.We | 1011 |
| abstract_inverted_index.Y. | 138, 625 |
| abstract_inverted_index.an | 170 |
| abstract_inverted_index.as | 316, 318, 430, 493, 495, 703 |
| abstract_inverted_index.be | 375, 675, 701, 774, 946, 995 |
| abstract_inverted_index.by | 74, 215, 765, 1121 |
| abstract_inverted_index.et | 25, 143, 181, 209, 448, 630, 825 |
| abstract_inverted_index.if | 915, 948 |
| abstract_inverted_index.in | 36, 87, 108, 134, 148, 173, 190, 243, 266, 289, 319, 377, 393, 426, 456, 479, 483, 490, 496, 527, 545, 559, 597, 601, 616, 635, 659, 677, 694, 744, 776, 794, 810, 815, 834, 851, 855, 865, 885, 919, 926, 932, 1005, 1019, 1029, 1046, 1062, 1111 |
| abstract_inverted_index.is | 3, 435, 951, 978 |
| abstract_inverted_index.it | 943 |
| abstract_inverted_index.of | 13, 17, 55, 105, 145, 184, 193, 211, 222, 322, 329, 367, 390, 412, 418, 423, 450, 532, 599, 612, 619, 632, 662, 670, 680, 760, 770, 783, 787, 807, 827, 881, 889, 896, 937, 970, 990, 1001, 1008, 1023, 1091, 1114 |
| abstract_inverted_index.or | 502, 578, 929, 962 |
| abstract_inverted_index.to | 95, 131, 168, 374, 432, 553, 576, 605, 738, 747, 755, 800, 859, 877, 913, 945, 953, 980, 994, 997, 1016, 1069, 1072, 1096, 1101, 1108 |
| abstract_inverted_index.(1) | 1095 |
| abstract_inverted_index.(2) | 1107 |
| abstract_inverted_index.16: | 839 |
| abstract_inverted_index.17: | 304 |
| abstract_inverted_index.18: | 199 |
| abstract_inverted_index.52: | 228 |
| abstract_inverted_index.61: | 117, 402 |
| abstract_inverted_index.Dis | 114, 399 |
| abstract_inverted_index.EC. | 712 |
| abstract_inverted_index.For | 49, 731 |
| abstract_inverted_index.Hep | 463, 1036 |
| abstract_inverted_index.Hou | 141, 628 |
| abstract_inverted_index.PDF | 260, 310, 471, 1131 |
| abstract_inverted_index.TLR | 788 |
| abstract_inverted_index.The | 162, 241, 358, 421, 571, 713, 803, 1054, 1089 |
| abstract_inverted_index.aim | 1090 |
| abstract_inverted_index.and | 11, 28, 33, 58, 83, 111, 187, 249, 268, 274, 291, 293, 333, 347, 380, 396, 453, 460, 488, 519, 561, 698, 729, 757, 792, 829, 868, 899, 935, 941, 959, 1026, 1033, 1060, 1080, 1086, 1106, 1118, 1124 |
| abstract_inverted_index.are | 70, 656, 863, 911, 1057 |
| abstract_inverted_index.but | 813 |
| abstract_inverted_index.can | 699, 869 |
| abstract_inverted_index.did | 564 |
| abstract_inverted_index.for | 67, 509, 535, 711, 874 |
| abstract_inverted_index.gut | 136, 146, 188, 299, 633, 883, 1003 |
| abstract_inverted_index.has | 63, 165, 371, 584, 735, 1043 |
| abstract_inverted_index.its | 59 |
| abstract_inverted_index.key | 592 |
| abstract_inverted_index.may | 674, 720, 773 |
| abstract_inverted_index.not | 71, 436, 565, 814 |
| abstract_inverted_index.one | 12 |
| abstract_inverted_index.rat | 781 |
| abstract_inverted_index.see | 914 |
| abstract_inverted_index.the | 4, 14, 50, 53, 84, 88, 96, 106, 135, 191, 250, 294, 298, 330, 348, 391, 410, 413, 416, 424, 474, 477, 480, 484, 511, 523, 538, 579, 595, 613, 617, 660, 678, 717, 816, 849, 852, 866, 879, 882, 890, 893, 917, 933, 949, 954, 964, 968, 999, 1002, 1006, 1017, 1020, 1047, 1051, 1063, 1103, 1112 |
| abstract_inverted_index.was | 594, 690, 797, 1094 |
| abstract_inverted_index.(16) | 159, 646 |
| abstract_inverted_index.(39) | 121, 406 |
| abstract_inverted_index.(54) | 843 |
| abstract_inverted_index.(90) | 355 |
| abstract_inverted_index.(EC) | 2 |
| abstract_inverted_index.(GI) | 91 |
| abstract_inverted_index.114: | 338 |
| abstract_inverted_index.136: | 42 |
| abstract_inverted_index.146: | 254 |
| abstract_inverted_index.150: | 155, 642 |
| abstract_inverted_index.348: | 351 |
| abstract_inverted_index.444: | 278 |
| abstract_inverted_index.A.D. | 236 |
| abstract_inverted_index.A.H. | 820 |
| abstract_inverted_index.Adv. | 464 |
| abstract_inverted_index.D.E. | 100, 385 |
| abstract_inverted_index.E.J. | 98, 383 |
| abstract_inverted_index.Full | 256, 258, 306, 308, 467, 469 |
| abstract_inverted_index.G.S. | 272 |
| abstract_inverted_index.Host | 301 |
| abstract_inverted_index.Iran | 196 |
| abstract_inverted_index.J.A. | 102, 387 |
| abstract_inverted_index.J.J. | 326 |
| abstract_inverted_index.K.E. | 285 |
| abstract_inverted_index.L.A. | 822 |
| abstract_inverted_index.M.R. | 175 |
| abstract_inverted_index.Med. | 197 |
| abstract_inverted_index.Open | 1132 |
| abstract_inverted_index.R.E. | 824 |
| abstract_inverted_index.R.J. | 238 |
| abstract_inverted_index.S.V. | 287 |
| abstract_inverted_index.Sci. | 115, 400 |
| abstract_inverted_index.Tang | 139, 626 |
| abstract_inverted_index.Text | 257, 259, 307, 309, 468, 470 |
| abstract_inverted_index.They | 514 |
| abstract_inverted_index.This | 751, 846 |
| abstract_inverted_index.W.S. | 345 |
| abstract_inverted_index.also | 758 |
| abstract_inverted_index.and, | 707 |
| abstract_inverted_index.been | 64, 166, 372, 736, 1067 |
| abstract_inverted_index.coli | 809 |
| abstract_inverted_index.diet | 1081 |
| abstract_inverted_index.done | 996 |
| abstract_inverted_index.from | 219, 548 |
| abstract_inverted_index.have | 132, 651, 923, 1066 |
| abstract_inverted_index.more | 657, 692 |
| abstract_inverted_index.most | 6 |
| abstract_inverted_index.need | 993 |
| abstract_inverted_index.oral | 368 |
| abstract_inverted_index.over | 1050 |
| abstract_inverted_index.part | 172 |
| abstract_inverted_index.past | 51, 1052 |
| abstract_inverted_index.play | 169 |
| abstract_inverted_index.risk | 77, 930 |
| abstract_inverted_index.role | 104, 183, 389, 422, 611, 880, 925 |
| abstract_inverted_index.taxa | 654 |
| abstract_inverted_index.that | 69, 574, 655, 673, 688, 716 |
| abstract_inverted_index.this | 440, 920, 1013, 1092 |
| abstract_inverted_index.tool | 873 |
| abstract_inverted_index.used | 702 |
| abstract_inverted_index.well | 317, 437, 494 |
| abstract_inverted_index.will | 922, 992 |
| abstract_inverted_index.with | 129, 150, 224, 297, 415, 498, 529, 588, 637, 664, 696, 723, 741, 906, 1126 |
| abstract_inverted_index.(109) | 342 |
| abstract_inverted_index.(189) | 232 |
| abstract_inverted_index.(238) | 313 |
| abstract_inverted_index.(EAC) | 1041 |
| abstract_inverted_index.2003; | 227 |
| abstract_inverted_index.2006; | 277 |
| abstract_inverted_index.2014; | 253 |
| abstract_inverted_index.2015; | 41, 198, 303, 350 |
| abstract_inverted_index.2016; | 116, 337, 401, 838 |
| abstract_inverted_index.2021; | 154, 641 |
| abstract_inverted_index.2022; | 465 |
| abstract_inverted_index.Ishaq | 176 |
| abstract_inverted_index.Kelly | 821 |
| abstract_inverted_index.Kreft | 823 |
| abstract_inverted_index.Lynch | 286 |
| abstract_inverted_index.TLR-4 | 771 |
| abstract_inverted_index.Using | 522 |
| abstract_inverted_index.among | 685 |
| abstract_inverted_index.bowel | 245 |
| abstract_inverted_index.cells | 725, 743 |
| abstract_inverted_index.colon | 745, 749 |
| abstract_inverted_index.could | 93, 1082 |
| abstract_inverted_index.fecal | 562, 580, 1119 |
| abstract_inverted_index.feces | 489, 661 |
| abstract_inverted_index.forms | 321 |
| abstract_inverted_index.found | 127, 373, 652, 691 |
| abstract_inverted_index.human | 331 |
| abstract_inverted_index.large | 988 |
| abstract_inverted_index.major | 34 |
| abstract_inverted_index.model | 782 |
| abstract_inverted_index.noted | 798, 955 |
| abstract_inverted_index.novel | 507 |
| abstract_inverted_index.other | 269, 606, 623, 686, 974 |
| abstract_inverted_index.risen | 1044 |
| abstract_inverted_index.shift | 544 |
| abstract_inverted_index.shown | 167, 737 |
| abstract_inverted_index.small | 894 |
| abstract_inverted_index.state | 1123 |
| abstract_inverted_index.study | 126, 847, 867, 891, 977, 1093 |
| abstract_inverted_index.tight | 586 |
| abstract_inverted_index.total | 556 |
| abstract_inverted_index.tract | 92 |
| abstract_inverted_index.tumor | 811 |
| abstract_inverted_index.upper | 89 |
| abstract_inverted_index.using | 505 |
| abstract_inverted_index.which | 370 |
| abstract_inverted_index.(1025) | 263 |
| abstract_inverted_index.(5975) | 282 |
| abstract_inverted_index.1Issue | 1038 |
| abstract_inverted_index.1–3, | 789 |
| abstract_inverted_index.Abrams | 101, 386 |
| abstract_inverted_index.Access | 1133 |
| abstract_inverted_index.Cancer | 346 |
| abstract_inverted_index.Fecal, | 1024 |
| abstract_inverted_index.Gevers | 239 |
| abstract_inverted_index.Google | 47, 122, 160, 201, 233, 264, 283, 314, 343, 356, 407, 472, 647, 844 |
| abstract_inverted_index.Larger | 909 |
| abstract_inverted_index.PubMed | 44, 119, 157, 230, 261, 280, 311, 340, 353, 404, 644, 841 |
| abstract_inverted_index.Scopus | 45, 120, 158, 231, 262, 281, 312, 341, 354, 405, 645, 842 |
| abstract_inverted_index.Tissue | 1027 |
| abstract_inverted_index.Xavier | 237 |
| abstract_inverted_index.across | 568 |
| abstract_inverted_index.al.The | 182 |
| abstract_inverted_index.asthma | 292 |
| abstract_inverted_index.cancer | 1, 8, 746 |
| abstract_inverted_index.causal | 983 |
| abstract_inverted_index.celiac | 194 |
| abstract_inverted_index.common | 7 |
| abstract_inverted_index.differ | 566 |
| abstract_inverted_index.direct | 709 |
| abstract_inverted_index.distal | 359 |
| abstract_inverted_index.eighth | 5 |
| abstract_inverted_index.favors | 967 |
| abstract_inverted_index.fecal, | 451 |
| abstract_inverted_index.field. | 1021 |
| abstract_inverted_index.future | 251 |
| abstract_inverted_index.having | 666 |
| abstract_inverted_index.method | 508, 1100 |
| abstract_inverted_index.mucosa | 221, 966 |
| abstract_inverted_index.needed | 912, 979 |
| abstract_inverted_index.normal | 801, 818 |
| abstract_inverted_index.number | 669, 895 |
| abstract_inverted_index.recent | 125 |
| abstract_inverted_index.reflux | 80, 381 |
| abstract_inverted_index.saliva | 577 |
| abstract_inverted_index.shifts | 1061 |
| abstract_inverted_index.status | 248 |
| abstract_inverted_index.tissue | 454, 533, 546, 796 |
| abstract_inverted_index.trying | 876 |
| abstract_inverted_index.unique | 363, 542 |
| abstract_inverted_index.useful | 872 |
| abstract_inverted_index.varium | 217 |
| abstract_inverted_index.words, | 975 |
| abstract_inverted_index.(20333) | 46 |
| abstract_inverted_index.(TLR-4) | 764 |
| abstract_inverted_index.5Ohkusa | 203 |
| abstract_inverted_index.6Kostic | 235 |
| abstract_inverted_index.Barrett | 457 |
| abstract_inverted_index.Cancer. | 40, 336, 837 |
| abstract_inverted_index.Dikshit | 23 |
| abstract_inverted_index.Dulaimi | 179 |
| abstract_inverted_index.Further | 682, 985 |
| abstract_inverted_index.Goedert | 325 |
| abstract_inverted_index.Methods | 858 |
| abstract_inverted_index.Metwaly | 444 |
| abstract_inverted_index.Ogihara | 207 |
| abstract_inverted_index.Okayasu | 205 |
| abstract_inverted_index.PAXgene | 524 |
| abstract_inverted_index.Pathog. | 153, 640 |
| abstract_inverted_index.Scholar | 48, 123, 161, 265, 315, 357, 408, 473, 648, 845 |
| abstract_inverted_index.Western | 1048 |
| abstract_inverted_index.allergy | 290 |
| abstract_inverted_index.altered | 376 |
| abstract_inverted_index.article | 1014 |
| abstract_inverted_index.authors | 475, 539, 572, 714, 804 |
| abstract_inverted_index.binding | 754 |
| abstract_inverted_index.cancer. | 750 |
| abstract_inverted_index.changes | 478, 958 |
| abstract_inverted_index.chronic | 907 |
| abstract_inverted_index.clarify | 981 |
| abstract_inverted_index.cohorts | 989 |
| abstract_inverted_index.colitis | 214 |
| abstract_inverted_index.colonic | 220 |
| abstract_inverted_index.control | 904 |
| abstract_inverted_index.current | 247 |
| abstract_inverted_index.disease | 428, 569, 1073, 1087, 1122 |
| abstract_inverted_index.drivers | 16 |
| abstract_inverted_index.enhance | 748 |
| abstract_inverted_index.factors | 1076 |
| abstract_inverted_index.fashion | 921 |
| abstract_inverted_index.finding | 593 |
| abstract_inverted_index.further | 976 |
| abstract_inverted_index.greater | 668 |
| abstract_inverted_index.growing | 66 |
| abstract_inverted_index.harmful | 671 |
| abstract_inverted_index.healthy | 491 |
| abstract_inverted_index.include | 892 |
| abstract_inverted_index.leading | 952 |
| abstract_inverted_index.methods | 32 |
| abstract_inverted_index.mucosal | 433, 724, 742, 957 |
| abstract_inverted_index.pathway | 833 |
| abstract_inverted_index.present | 86 |
| abstract_inverted_index.primary | 15 |
| abstract_inverted_index.provide | 870 |
| abstract_inverted_index.rapidly | 65 |
| abstract_inverted_index.reasons | 68, 1056 |
| abstract_inverted_index.related | 431 |
| abstract_inverted_index.remains | 944 |
| abstract_inverted_index.saliva, | 485, 1115 |
| abstract_inverted_index.samples | 563, 897, 1120 |
| abstract_inverted_index.several | 320 |
| abstract_inverted_index.similar | 768 |
| abstract_inverted_index.species | 600 |
| abstract_inverted_index.studies | 328, 910, 986 |
| abstract_inverted_index.surface | 727 |
| abstract_inverted_index.through | 726 |
| abstract_inverted_index.tissue, | 487, 1117 |
| abstract_inverted_index.tissues | 812 |
| abstract_inverted_index.whether | 963 |
| abstract_inverted_index.2012.Int | 38 |
| abstract_inverted_index.Abstract | 466 |
| abstract_inverted_index.Analysis | 1022 |
| abstract_inverted_index.Finally, | 940 |
| abstract_inverted_index.GLOBOCAN | 37 |
| abstract_inverted_index.However, | 555, 622 |
| abstract_inverted_index.Microbe. | 302 |
| abstract_inverted_index.Multiple | 1075 |
| abstract_inverted_index.Scholar, | 202, 234 |
| abstract_inverted_index.abnormal | 965 |
| abstract_inverted_index.abundant | 658 |
| abstract_inverted_index.adjacent | 817 |
| abstract_inverted_index.analysis | 683 |
| abstract_inverted_index.analyzed | 898 |
| abstract_inverted_index.bacteria | 672 |
| abstract_inverted_index.biopsies | 534, 547 |
| abstract_inverted_index.clinical | 927 |
| abstract_inverted_index.cohorts. | 1129 |
| abstract_inverted_index.compared | 552, 575, 604, 799 |
| abstract_inverted_index.composed | 365 |
| abstract_inverted_index.contains | 361 |
| abstract_inverted_index.decades. | 1053 |
| abstract_inverted_index.defined. | 438 |
| abstract_inverted_index.detected | 805 |
| abstract_inverted_index.directly | 722, 740 |
| abstract_inverted_index.disease, | 81 |
| abstract_inverted_index.disease: | 246 |
| abstract_inverted_index.emerging | 295 |
| abstract_inverted_index.example, | 732 |
| abstract_inverted_index.external | 1128 |
| abstract_inverted_index.factors. | 78 |
| abstract_inverted_index.feasible | 1099 |
| abstract_inverted_index.fixation | 525 |
| abstract_inverted_index.globally | 10 |
| abstract_inverted_index.identify | 1097 |
| abstract_inverted_index.increase | 1070 |
| abstract_inverted_index.interact | 721, 739 |
| abstract_inverted_index.involved | 676, 775 |
| abstract_inverted_index.involves | 753 |
| abstract_inverted_index.isolated | 218 |
| abstract_inverted_index.obesity, | 82 |
| abstract_inverted_index.paraffin | 530 |
| abstract_inverted_index.patients | 128, 149, 223, 492, 497, 551, 636, 695, 905, 991 |
| abstract_inverted_index.patterns | 35 |
| abstract_inverted_index.possibly | 700 |
| abstract_inverted_index.properly | 72 |
| abstract_inverted_index.proteins | 728 |
| abstract_inverted_index.recently | 650 |
| abstract_inverted_index.receptor | 762, 831 |
| abstract_inverted_index.richness | 598 |
| abstract_inverted_index.salivary | 452, 560 |
| abstract_inverted_index.sources, | 31 |
| abstract_inverted_index.specific | 886, 971 |
| abstract_inverted_index.validate | 1125 |
| abstract_inverted_index.Barrett's | 61, 109, 378, 394, 500, 1030 |
| abstract_inverted_index.Freedberg | 99, 384 |
| abstract_inverted_index.Full-Text | 1130 |
| abstract_inverted_index.Potential | 103, 388 |
| abstract_inverted_index.Reitmeier | 446 |
| abstract_inverted_index.Salivary, | 1025 |
| abstract_inverted_index.abundance | 806 |
| abstract_inverted_index.al.Cancer | 26 |
| abstract_inverted_index.assessing | 916 |
| abstract_inverted_index.biomarker | 706 |
| abstract_inverted_index.cancer.Br | 334 |
| abstract_inverted_index.cancerous | 795 |
| abstract_inverted_index.controls. | 554 |
| abstract_inverted_index.countries | 1049 |
| abstract_inverted_index.currently | 75 |
| abstract_inverted_index.described | 864 |
| abstract_inverted_index.diagnosed | 9 |
| abstract_inverted_index.diagnosis | 934 |
| abstract_inverted_index.diseases. | 887 |
| abstract_inverted_index.diversity | 558 |
| abstract_inverted_index.dysbiosis | 133, 950 |
| abstract_inverted_index.dysplasia | 434, 459 |
| abstract_inverted_index.embedding | 531 |
| abstract_inverted_index.esophagus | 110, 360, 379, 395 |
| abstract_inverted_index.essential | 171 |
| abstract_inverted_index.explained | 73 |
| abstract_inverted_index.incidence | 27, 54, 1042 |
| abstract_inverted_index.including | 1077 |
| abstract_inverted_index.influence | 1083 |
| abstract_inverted_index.involving | 987 |
| abstract_inverted_index.mechanism | 769 |
| abstract_inverted_index.mediators | 186 |
| abstract_inverted_index.metabolic | 275 |
| abstract_inverted_index.microbes. | 972 |
| abstract_inverted_index.microbial | 543, 557, 705 |
| abstract_inverted_index.mortality | 29 |
| abstract_inverted_index.nucleatum | 734 |
| abstract_inverted_index.patients, | 663 |
| abstract_inverted_index.performed | 515 |
| abstract_inverted_index.possibly, | 708 |
| abstract_inverted_index.potential | 610, 901 |
| abstract_inverted_index.primarily | 366 |
| abstract_inverted_index.published | 649 |
| abstract_inverted_index.reduction | 596 |
| abstract_inverted_index.revealed, | 684 |
| abstract_inverted_index.signaling | 832 |
| abstract_inverted_index.suggested | 573, 715 |
| abstract_inverted_index.technique | 526 |
| abstract_inverted_index.toll-like | 761, 830 |
| abstract_inverted_index.treatment | 710 |
| abstract_inverted_index.utilizing | 903 |
| abstract_inverted_index.52Crossref | 840 |
| abstract_inverted_index.Dysplasia, | 1032 |
| abstract_inverted_index.E-cadherin | 756 |
| abstract_inverted_index.Esophageal | 0, 1034 |
| abstract_inverted_index.Esophagus, | 1031 |
| abstract_inverted_index.Microbiome | 1028 |
| abstract_inverted_index.Microbiota | 288 |
| abstract_inverted_index.abundantly | 693 |
| abstract_inverted_index.activation | 759, 772 |
| abstract_inverted_index.assessment | 931 |
| abstract_inverted_index.bacterium, | 687 |
| abstract_inverted_index.carcinoma. | 681 |
| abstract_inverted_index.confounder | 902 |
| abstract_inverted_index.contribute | 94 |
| abstract_inverted_index.determined | 947 |
| abstract_inverted_index.disorders. | 420 |
| abstract_inverted_index.dysplasia, | 499 |
| abstract_inverted_index.esophageal | 56, 112, 151, 397, 419, 427, 461, 486, 503, 549, 589, 602, 620, 638, 777, 784, 835, 856, 938, 956, 1009, 1064, 1116 |
| abstract_inverted_index.esophagus, | 62, 458, 501 |
| abstract_inverted_index.excitement | 1018 |
| abstract_inverted_index.gastritis. | 908 |
| abstract_inverted_index.highlights | 848 |
| abstract_inverted_index.infectious | 185 |
| abstract_inverted_index.integrated | 517 |
| abstract_inverted_index.intestinal | 270 |
| abstract_inverted_index.lifestyle, | 1079 |
| abstract_inverted_index.management | 936 |
| abstract_inverted_index.microbiome | 85, 107, 164, 189, 242, 332, 364, 392, 414, 425, 455, 482, 536, 583, 615, 719, 854, 862, 884, 918, 1004, 1065, 1084, 1113 |
| abstract_inverted_index.microbiota | 147, 634, 828 |
| abstract_inverted_index.postulated | 1068 |
| abstract_inverted_index.precursor, | 60 |
| abstract_inverted_index.subsequent | 960 |
| abstract_inverted_index.suggesting | 608 |
| abstract_inverted_index.ulcerative | 213, 225 |
| abstract_inverted_index.underlying | 1055 |
| abstract_inverted_index.understand | 878, 998 |
| abstract_inverted_index.worldwide: | 30 |
| abstract_inverted_index.Escherichia | 808 |
| abstract_inverted_index.Lachnospira | 689 |
| abstract_inverted_index.Limitations | 888 |
| abstract_inverted_index.acknowledge | 1012 |
| abstract_inverted_index.al.Analysis | 449 |
| abstract_inverted_index.association | 411, 587 |
| abstract_inverted_index.composition | 1085 |
| abstract_inverted_index.conjunction | 528 |
| abstract_inverted_index.contributes | 1015 |
| abstract_inverted_index.differences | 850, 1110 |
| abstract_inverted_index.epithelium. | 802 |
| abstract_inverted_index.established | 76 |
| abstract_inverted_index.interaction | 752 |
| abstract_inverted_index.investigate | 1109 |
| abstract_inverted_index.microbiome, | 581, 1105 |
| abstract_inverted_index.microbiome. | 513 |
| abstract_inverted_index.microbiota, | 369 |
| abstract_inverted_index.phenotypes. | 607 |
| abstract_inverted_index.progression | 429, 679 |
| abstract_inverted_index.standardize | 860 |
| abstract_inverted_index.understood, | 1059 |
| abstract_inverted_index.AdvancesVol. | 1037 |
| abstract_inverted_index.Inflammation | 273 |
| abstract_inverted_index.al.Dysbiosis | 144, 631 |
| abstract_inverted_index.al.Induction | 210 |
| abstract_inverted_index.characterize | 1102 |
| abstract_inverted_index.colitis.Gut. | 226 |
| abstract_inverted_index.colonization | 969 |
| abstract_inverted_index.contribution | 1000 |
| abstract_inverted_index.demonstrated | 540 |
| abstract_inverted_index.development. | 1074 |
| abstract_inverted_index.disease.Arch | 195 |
| abstract_inverted_index.dramatically | 1045 |
| abstract_inverted_index.establishing | 409 |
| abstract_inverted_index.experimental | 212 |
| abstract_inverted_index.importantly, | 942 |
| abstract_inverted_index.incompletely | 1058 |
| abstract_inverted_index.inflammatory | 244 |
| abstract_inverted_index.investigated | 476 |
| abstract_inverted_index.medications, | 1078 |
| abstract_inverted_index.metabolites. | 730 |
| abstract_inverted_index.pathogenesis | 192, 618, 1007 |
| abstract_inverted_index.progression. | 570, 1088 |
| abstract_inverted_index.relationship | 296 |
| abstract_inverted_index.simultaneous | 516 |
| abstract_inverted_index.studies3Deng | 624 |
| abstract_inverted_index.upregulation | 786 |
| abstract_inverted_index.244-249PubMed | 200 |
| abstract_inverted_index.79-83Crossref | 229 |
| abstract_inverted_index.80-86Crossref | 352 |
| abstract_inverted_index.Epidemiologic | 327 |
| abstract_inverted_index.Fusobacterium | 216, 733 |
| abstract_inverted_index.Soerjomataram | 21 |
| abstract_inverted_index.cancer.Microb | 152, 639 |
| abstract_inverted_index.correlations. | 521 |
| abstract_inverted_index.half-century, | 52 |
| abstract_inverted_index.health,4Nejad | 174 |
| abstract_inverted_index.investigators | 875 |
| abstract_inverted_index.relationship. | 984 |
| abstract_inverted_index.significantly | 567 |
| abstract_inverted_index.104709Crossref | 156, 643 |
| abstract_inverted_index.adenocarcinoma | 57, 504, 550, 603, 961, 1040 |
| abstract_inverted_index.cancer-related | 18 |
| abstract_inverted_index.characterizing | 510 |
| abstract_inverted_index.deaths.1Ferlay | 19 |
| abstract_inverted_index.investigation, | 537 |
| abstract_inverted_index.predisposition | 1071 |
| abstract_inverted_index.study,11Radani | 442 |
| abstract_inverted_index.upper-GI-tract | 481 |
| abstract_inverted_index.237-242Crossref | 339 |
| abstract_inverted_index.592-602Abstract | 305 |
| abstract_inverted_index.860-867Crossref | 279 |
| abstract_inverted_index.adenocarcinoma, | 785 |
| abstract_inverted_index.adenocarcinoma. | 590, 621, 857, 939, 1010 |
| abstract_inverted_index.al.Associations | 826 |
| abstract_inverted_index.carcinogenesis. | 778 |
| abstract_inverted_index.cross-sectional | 441 |
| abstract_inverted_index.decision-making | 928 |
| abstract_inverted_index.epidemiological | 520 |
| abstract_inverted_index.microbiome.Cell | 300 |
| abstract_inverted_index.pathophysiology | 417 |
| abstract_inverted_index.Gastroesophageal | 79 |
| abstract_inverted_index.cancer.9Vogtmann | 323 |
| abstract_inverted_index.gastrointestinal | 90 |
| abstract_inverted_index.microbiome.3Deng | 137 |
| abstract_inverted_index.1489-1499Abstract | 255 |
| abstract_inverted_index.2217-2225Crossref | 118, 403 |
| abstract_inverted_index.E359-E386Crossref | 43 |
| abstract_inverted_index.Scholar,10Garrett | 344 |
| abstract_inverted_index.Scholar,8Fujimura | 284 |
| abstract_inverted_index.disorders.Nature. | 276 |
| abstract_inverted_index.mucosa-associated | 614, 718, 853, 861 |
| abstract_inverted_index.tissue-associated | 512, 1104 |
| abstract_inverted_index.5PreviewEsophageal | 1039 |
| abstract_inverted_index.adenocarcinoma.BMC | 836 |
| abstract_inverted_index.adenocarcinoma.Dig | 113, 398 |
| abstract_inverted_index.epithelium.12Zaidi | 819 |
| abstract_inverted_index.esophagitis,2Snider | 382 |
| abstract_inverted_index.microbiota.Science. | 349 |
| abstract_inverted_index.AdenocarcinomaGastro | 1035 |
| abstract_inverted_index.lipopolysaccharides. | 766 |
| abstract_inverted_index.pathogenesis.2Snider | 97 |
| abstract_inverted_index.tissue-biopsy-linked | 582 |
| abstract_inverted_index.adenocarcinoma.Gastro | 462 |
| abstract_inverted_index.clinical-pathological | 518 |
| abstract_inverted_index.ahead.Gastroenterology. | 252 |
| abstract_inverted_index.disorders,7Hotamisligil | 271 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5091361579 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 2 |
| corresponding_institution_ids | https://openalex.org/I150468666, https://openalex.org/I4210149289 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.8700000047683716 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.06105003 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |