Early-life stress exposure impacts the hippocampal synaptic proteome in a mouse model of Alzheimer’s disease: age- and pathology-dependent effects on mitochondrial proteins Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.04.20.537660
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1101/2023.04.20.537660
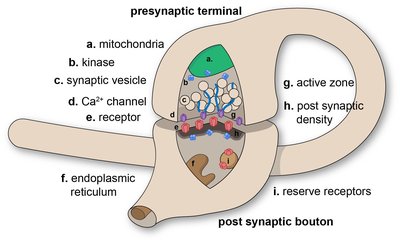
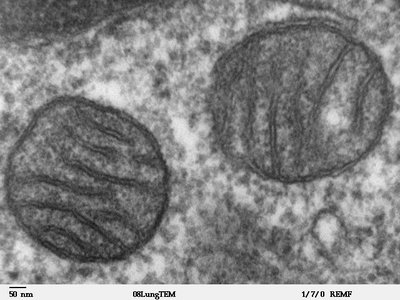
Epidemiological evidence indicates that early life stress (ES) exposure increases the risk for later-life diseases, such as Alzheimer’s disease (AD). Accordingly, we and others have shown that ES aggravates the development of, and response to, amyloid-beta (Aβ) pathology in animal models. Moreover, ES-exposed transgenic APP/PS1 mice display deficits in both cognitive flexibility and synaptic function. As the mechanisms behind these changes were unclear, we here investigated how exposure to ES, using the limited nesting and bedding model, affects the synaptic proteome across 2 different ages in both wildtype and APP/PS1 transgenic mice. We found that, compared to wildtype mice, the hippocampal synaptosomes of APP/PS1 mice at an early pathological stage (4 months) showed a higher abundance of mitochondrial proteins and lower levels of proteins involved in actin dynamics. Interestingly, ES exposure in wildtype mice had similar effects on the level of mitochondrial and actin-related synaptosomal proteins at this age, whereas ES exposure had no additional effect on the synaptosomal proteome of early-stage APP/PS1 mice. Accordingly, ultrastructural analysis of the synapse using electron microscopy in a follow-up cohort showed fewer mitochondria in pre- and post-synaptic compartments of APP/PS1 and ES-exposed mice, respectively. At a later pathological stage (10 months), the hippocampal synaptic proteome of APP/PS1 mice revealed an upregulation of proteins related to Aβ processing, that was accompanied by a downregulation of proteins related to postsynaptic receptor endocytosis. ES exposure no longer affected the synaptic proteome of wildtype animals by this age, whereas it affected the expression of astrocytic proteins involved in lipid metabolism in APP/PS1 mice. We confirmed a dysregulation of astrocyte protein expression in a separate cohort of 12-month-old mice, by immunostaining for the alpha subunit of the mitochondrial trifunctional protein and fatty acid synthase in astrocytes. In conclusion, our data suggest that ES and amyloidosis share pathogenic pathways involving synaptic mitochondrial dysfunction and astrocytic lipid metabolism. These pathways might be underlying contributors to the long-term aggravation of the APP/PS1 phenotype by ES, as well as to the ES-associated risk for AD progression. These data are publicly accessible online as a web app via https://amsterdamstudygroup.shinyapps.io/ES_Synaptosome_Proteomics_Visualizer/ .
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2023.04.20.537660
- https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdf
- OA Status
- green
- References
- 128
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4366550673
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4366550673Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2023.04.20.537660Digital Object Identifier
- Title
-
Early-life stress exposure impacts the hippocampal synaptic proteome in a mouse model of Alzheimer’s disease: age- and pathology-dependent effects on mitochondrial proteinsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-04-20Full publication date if available
- Authors
-
Janssen M. Kotah, Mandy S. J. Kater, Lianne Hoeijmakers, Niek Brosens, Sylvie L. Lesuis, Roberta Tandari, Luca Marchetto, Ella Yusaf, August B. Smit, Paul J. Lucassen, Harm J. Krugers, Mark H. G. Verheijen, Anikó KőrösiList of authors in order
- Landing page
-
https://doi.org/10.1101/2023.04.20.537660Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdfDirect OA link when available
- Concepts
-
Proteome, Downregulation and upregulation, Hippocampal formation, Genetically modified mouse, Synapse, Biology, Neurodegeneration, Mitochondrion, Cell biology, Endocrinology, Neuroscience, Internal medicine, Transgene, Disease, Medicine, Bioinformatics, Biochemistry, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
128Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4366550673 |
|---|---|
| doi | https://doi.org/10.1101/2023.04.20.537660 |
| ids.doi | https://doi.org/10.1101/2023.04.20.537660 |
| ids.openalex | https://openalex.org/W4366550673 |
| fwci | |
| type | preprint |
| title | Early-life stress exposure impacts the hippocampal synaptic proteome in a mouse model of Alzheimer’s disease: age- and pathology-dependent effects on mitochondrial proteins |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10086 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9995999932289124 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2737 |
| topics[0].subfield.display_name | Physiology |
| topics[0].display_name | Alzheimer's disease research and treatments |
| topics[1].id | https://openalex.org/T11337 |
| topics[1].field.id | https://openalex.org/fields/28 |
| topics[1].field.display_name | Neuroscience |
| topics[1].score | 0.9994999766349792 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2803 |
| topics[1].subfield.display_name | Biological Psychiatry |
| topics[1].display_name | Tryptophan and brain disorders |
| topics[2].id | https://openalex.org/T10529 |
| topics[2].field.id | https://openalex.org/fields/28 |
| topics[2].field.display_name | Neuroscience |
| topics[2].score | 0.9980000257492065 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/2802 |
| topics[2].subfield.display_name | Behavioral Neuroscience |
| topics[2].display_name | Stress Responses and Cortisol |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C104397665 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7061899900436401 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q860947 |
| concepts[0].display_name | Proteome |
| concepts[1].id | https://openalex.org/C127561419 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6501414775848389 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q224527 |
| concepts[1].display_name | Downregulation and upregulation |
| concepts[2].id | https://openalex.org/C148762608 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6021726131439209 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q3748185 |
| concepts[2].display_name | Hippocampal formation |
| concepts[3].id | https://openalex.org/C141035611 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5662539601325989 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q5532934 |
| concepts[3].display_name | Genetically modified mouse |
| concepts[4].id | https://openalex.org/C127445978 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5553134679794312 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q187181 |
| concepts[4].display_name | Synapse |
| concepts[5].id | https://openalex.org/C86803240 |
| concepts[5].level | 0 |
| concepts[5].score | 0.5356540083885193 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[5].display_name | Biology |
| concepts[6].id | https://openalex.org/C2776925932 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4681226313114166 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1755122 |
| concepts[6].display_name | Neurodegeneration |
| concepts[7].id | https://openalex.org/C28859421 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4365765452384949 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q39572 |
| concepts[7].display_name | Mitochondrion |
| concepts[8].id | https://openalex.org/C95444343 |
| concepts[8].level | 1 |
| concepts[8].score | 0.3903723657131195 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[8].display_name | Cell biology |
| concepts[9].id | https://openalex.org/C134018914 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3359279930591583 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q162606 |
| concepts[9].display_name | Endocrinology |
| concepts[10].id | https://openalex.org/C169760540 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3201944828033447 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q207011 |
| concepts[10].display_name | Neuroscience |
| concepts[11].id | https://openalex.org/C126322002 |
| concepts[11].level | 1 |
| concepts[11].score | 0.3051658570766449 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[11].display_name | Internal medicine |
| concepts[12].id | https://openalex.org/C102230213 |
| concepts[12].level | 3 |
| concepts[12].score | 0.29052186012268066 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q430117 |
| concepts[12].display_name | Transgene |
| concepts[13].id | https://openalex.org/C2779134260 |
| concepts[13].level | 2 |
| concepts[13].score | 0.1988827884197235 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q12136 |
| concepts[13].display_name | Disease |
| concepts[14].id | https://openalex.org/C71924100 |
| concepts[14].level | 0 |
| concepts[14].score | 0.18692246079444885 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[14].display_name | Medicine |
| concepts[15].id | https://openalex.org/C60644358 |
| concepts[15].level | 1 |
| concepts[15].score | 0.12827977538108826 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q128570 |
| concepts[15].display_name | Bioinformatics |
| concepts[16].id | https://openalex.org/C55493867 |
| concepts[16].level | 1 |
| concepts[16].score | 0.12258809804916382 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[16].display_name | Biochemistry |
| concepts[17].id | https://openalex.org/C104317684 |
| concepts[17].level | 2 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[17].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/proteome |
| keywords[0].score | 0.7061899900436401 |
| keywords[0].display_name | Proteome |
| keywords[1].id | https://openalex.org/keywords/downregulation-and-upregulation |
| keywords[1].score | 0.6501414775848389 |
| keywords[1].display_name | Downregulation and upregulation |
| keywords[2].id | https://openalex.org/keywords/hippocampal-formation |
| keywords[2].score | 0.6021726131439209 |
| keywords[2].display_name | Hippocampal formation |
| keywords[3].id | https://openalex.org/keywords/genetically-modified-mouse |
| keywords[3].score | 0.5662539601325989 |
| keywords[3].display_name | Genetically modified mouse |
| keywords[4].id | https://openalex.org/keywords/synapse |
| keywords[4].score | 0.5553134679794312 |
| keywords[4].display_name | Synapse |
| keywords[5].id | https://openalex.org/keywords/biology |
| keywords[5].score | 0.5356540083885193 |
| keywords[5].display_name | Biology |
| keywords[6].id | https://openalex.org/keywords/neurodegeneration |
| keywords[6].score | 0.4681226313114166 |
| keywords[6].display_name | Neurodegeneration |
| keywords[7].id | https://openalex.org/keywords/mitochondrion |
| keywords[7].score | 0.4365765452384949 |
| keywords[7].display_name | Mitochondrion |
| keywords[8].id | https://openalex.org/keywords/cell-biology |
| keywords[8].score | 0.3903723657131195 |
| keywords[8].display_name | Cell biology |
| keywords[9].id | https://openalex.org/keywords/endocrinology |
| keywords[9].score | 0.3359279930591583 |
| keywords[9].display_name | Endocrinology |
| keywords[10].id | https://openalex.org/keywords/neuroscience |
| keywords[10].score | 0.3201944828033447 |
| keywords[10].display_name | Neuroscience |
| keywords[11].id | https://openalex.org/keywords/internal-medicine |
| keywords[11].score | 0.3051658570766449 |
| keywords[11].display_name | Internal medicine |
| keywords[12].id | https://openalex.org/keywords/transgene |
| keywords[12].score | 0.29052186012268066 |
| keywords[12].display_name | Transgene |
| keywords[13].id | https://openalex.org/keywords/disease |
| keywords[13].score | 0.1988827884197235 |
| keywords[13].display_name | Disease |
| keywords[14].id | https://openalex.org/keywords/medicine |
| keywords[14].score | 0.18692246079444885 |
| keywords[14].display_name | Medicine |
| keywords[15].id | https://openalex.org/keywords/bioinformatics |
| keywords[15].score | 0.12827977538108826 |
| keywords[15].display_name | Bioinformatics |
| keywords[16].id | https://openalex.org/keywords/biochemistry |
| keywords[16].score | 0.12258809804916382 |
| keywords[16].display_name | Biochemistry |
| language | en |
| locations[0].id | doi:10.1101/2023.04.20.537660 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc-nd |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc-nd |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2023.04.20.537660 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5065323781 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-3005-730X |
| authorships[0].author.display_name | Janssen M. Kotah |
| authorships[0].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Janssen M. Kotah |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[1].author.id | https://openalex.org/A5062636371 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-4832-2597 |
| authorships[1].author.display_name | Mandy S. J. Kater |
| authorships[1].countries | NL |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210108594, https://openalex.org/I865915315 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands |
| authorships[1].institutions[0].id | https://openalex.org/I4210108594 |
| authorships[1].institutions[0].ror | https://ror.org/01x2d9f70 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I4210108594 |
| authorships[1].institutions[0].country_code | NL |
| authorships[1].institutions[0].display_name | Amsterdam Neuroscience |
| authorships[1].institutions[1].id | https://openalex.org/I865915315 |
| authorships[1].institutions[1].ror | https://ror.org/008xxew50 |
| authorships[1].institutions[1].type | education |
| authorships[1].institutions[1].lineage | https://openalex.org/I865915315 |
| authorships[1].institutions[1].country_code | NL |
| authorships[1].institutions[1].display_name | Vrije Universiteit Amsterdam |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Mandy S.J. Kater |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands |
| authorships[2].author.id | https://openalex.org/A5068459181 |
| authorships[2].author.orcid | https://orcid.org/0009-0004-7999-1886 |
| authorships[2].author.display_name | Lianne Hoeijmakers |
| authorships[2].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Lianne Hoeijmakers |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[3].author.id | https://openalex.org/A5008733090 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-8347-8522 |
| authorships[3].author.display_name | Niek Brosens |
| authorships[3].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Niek Brosens |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[4].author.id | https://openalex.org/A5061052429 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-7727-9271 |
| authorships[4].author.display_name | Sylvie L. Lesuis |
| authorships[4].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Sylvie L. Lesuis |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[5].author.id | https://openalex.org/A5044664924 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Roberta Tandari |
| authorships[5].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Roberta Tandari |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[6].author.id | https://openalex.org/A5014567492 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-5678-3006 |
| authorships[6].author.display_name | Luca Marchetto |
| authorships[6].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Luca Marchetto |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[7].author.id | https://openalex.org/A5057050170 |
| authorships[7].author.orcid | |
| authorships[7].author.display_name | Ella Yusaf |
| authorships[7].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Ella Yusaf |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[8].author.id | https://openalex.org/A5046661343 |
| authorships[8].author.orcid | https://orcid.org/0000-0002-2286-1587 |
| authorships[8].author.display_name | August B. Smit |
| authorships[8].countries | NL |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I4210108594, https://openalex.org/I865915315 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands |
| authorships[8].institutions[0].id | https://openalex.org/I4210108594 |
| authorships[8].institutions[0].ror | https://ror.org/01x2d9f70 |
| authorships[8].institutions[0].type | facility |
| authorships[8].institutions[0].lineage | https://openalex.org/I4210108594 |
| authorships[8].institutions[0].country_code | NL |
| authorships[8].institutions[0].display_name | Amsterdam Neuroscience |
| authorships[8].institutions[1].id | https://openalex.org/I865915315 |
| authorships[8].institutions[1].ror | https://ror.org/008xxew50 |
| authorships[8].institutions[1].type | education |
| authorships[8].institutions[1].lineage | https://openalex.org/I865915315 |
| authorships[8].institutions[1].country_code | NL |
| authorships[8].institutions[1].display_name | Vrije Universiteit Amsterdam |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | August B. Smit |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands |
| authorships[9].author.id | https://openalex.org/A5000501122 |
| authorships[9].author.orcid | https://orcid.org/0000-0001-9708-9133 |
| authorships[9].author.display_name | Paul J. Lucassen |
| authorships[9].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Paul J. Lucassen |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[10].author.id | https://openalex.org/A5086284254 |
| authorships[10].author.orcid | https://orcid.org/0000-0002-7926-7001 |
| authorships[10].author.display_name | Harm J. Krugers |
| authorships[10].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Harm Krugers |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[11].author.id | https://openalex.org/A5027762242 |
| authorships[11].author.orcid | https://orcid.org/0000-0002-3739-3755 |
| authorships[11].author.display_name | Mark H. G. Verheijen |
| authorships[11].countries | NL |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I4210108594, https://openalex.org/I865915315 |
| authorships[11].affiliations[0].raw_affiliation_string | Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands |
| authorships[11].institutions[0].id | https://openalex.org/I4210108594 |
| authorships[11].institutions[0].ror | https://ror.org/01x2d9f70 |
| authorships[11].institutions[0].type | facility |
| authorships[11].institutions[0].lineage | https://openalex.org/I4210108594 |
| authorships[11].institutions[0].country_code | NL |
| authorships[11].institutions[0].display_name | Amsterdam Neuroscience |
| authorships[11].institutions[1].id | https://openalex.org/I865915315 |
| authorships[11].institutions[1].ror | https://ror.org/008xxew50 |
| authorships[11].institutions[1].type | education |
| authorships[11].institutions[1].lineage | https://openalex.org/I865915315 |
| authorships[11].institutions[1].country_code | NL |
| authorships[11].institutions[1].display_name | Vrije Universiteit Amsterdam |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Mark H.G. Verheijen |
| authorships[11].is_corresponding | False |
| authorships[11].raw_affiliation_strings | Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands |
| authorships[12].author.id | https://openalex.org/A5076907548 |
| authorships[12].author.orcid | https://orcid.org/0000-0001-9701-3331 |
| authorships[12].author.display_name | Anikó Kőrösi |
| authorships[12].affiliations[0].raw_affiliation_string | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| authorships[12].author_position | last |
| authorships[12].raw_author_name | Aniko Korosi |
| authorships[12].is_corresponding | True |
| authorships[12].raw_affiliation_strings | Brain Plasticity Group, Swammerdam Institute for Life Sciences – Center for Neuroscience, University of Amsterdam |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Early-life stress exposure impacts the hippocampal synaptic proteome in a mouse model of Alzheimer’s disease: age- and pathology-dependent effects on mitochondrial proteins |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10086 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9995999932289124 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2737 |
| primary_topic.subfield.display_name | Physiology |
| primary_topic.display_name | Alzheimer's disease research and treatments |
| related_works | https://openalex.org/W2794217300, https://openalex.org/W2764192351, https://openalex.org/W2965213848, https://openalex.org/W3031633328, https://openalex.org/W1886844647, https://openalex.org/W4313648027, https://openalex.org/W4323652623, https://openalex.org/W4327554146, https://openalex.org/W4308332022, https://openalex.org/W2903220633 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2023.04.20.537660 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc-nd |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2023.04.20.537660 |
| primary_location.id | doi:10.1101/2023.04.20.537660 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc-nd |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2023/04/20/2023.04.20.537660.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc-nd |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2023.04.20.537660 |
| publication_date | 2023-04-20 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W1999794509, https://openalex.org/W2026970282, https://openalex.org/W2104931471, https://openalex.org/W3164696834, https://openalex.org/W2156244858, https://openalex.org/W2018926899, https://openalex.org/W1987375729, https://openalex.org/W2164402789, https://openalex.org/W2344041198, https://openalex.org/W2884231443, https://openalex.org/W2009189397, https://openalex.org/W1964748520, https://openalex.org/W2155873686, https://openalex.org/W2065075723, https://openalex.org/W2077824607, https://openalex.org/W2526656473, https://openalex.org/W2791785701, https://openalex.org/W2791831747, https://openalex.org/W1975141413, https://openalex.org/W2142943752, https://openalex.org/W2586088462, https://openalex.org/W4306760649, https://openalex.org/W2151236915, https://openalex.org/W3176022318, https://openalex.org/W1972165428, https://openalex.org/W2111570868, https://openalex.org/W1988125482, https://openalex.org/W1620047260, https://openalex.org/W2566196214, https://openalex.org/W3012582992, https://openalex.org/W2802451112, https://openalex.org/W2959937585, https://openalex.org/W2911151802, https://openalex.org/W2915389159, https://openalex.org/W2020486984, https://openalex.org/W2015933608, https://openalex.org/W2168334242, https://openalex.org/W2086355707, https://openalex.org/W2025563134, https://openalex.org/W2147435466, https://openalex.org/W4200530045, https://openalex.org/W3173625082, https://openalex.org/W2792494265, https://openalex.org/W3008438024, https://openalex.org/W4306746333, https://openalex.org/W3125441563, https://openalex.org/W4312065586, https://openalex.org/W3198684761, https://openalex.org/W1979283544, https://openalex.org/W2794480084, https://openalex.org/W2948949781, https://openalex.org/W3106539179, https://openalex.org/W2007586038, https://openalex.org/W2018241116, https://openalex.org/W2009744263, https://openalex.org/W2112075758, https://openalex.org/W2012034410, https://openalex.org/W2163811645, https://openalex.org/W2885190910, https://openalex.org/W4212868491, https://openalex.org/W2762910600, https://openalex.org/W3033303495, https://openalex.org/W3118635735, https://openalex.org/W1983930192, https://openalex.org/W134802319, https://openalex.org/W2061387301, https://openalex.org/W3195477286, https://openalex.org/W2587549569, https://openalex.org/W3094724728, https://openalex.org/W3041386918, https://openalex.org/W2103024032, https://openalex.org/W2911248861, https://openalex.org/W4210538436, https://openalex.org/W2098548022, https://openalex.org/W2774690079, https://openalex.org/W2890398446, https://openalex.org/W3044915966, https://openalex.org/W2969562288, https://openalex.org/W1685999671, https://openalex.org/W4220830145, https://openalex.org/W2765088010, https://openalex.org/W4225854207, https://openalex.org/W3209041513, https://openalex.org/W4220870023, https://openalex.org/W2943862192, https://openalex.org/W2024797763, https://openalex.org/W3022167154, https://openalex.org/W3177336769, https://openalex.org/W2035815961, https://openalex.org/W1510830988, https://openalex.org/W2100621375, https://openalex.org/W2272690659, https://openalex.org/W1968674432, https://openalex.org/W1591312627, https://openalex.org/W1991902515, https://openalex.org/W2161685550, https://openalex.org/W1592529377, https://openalex.org/W3164253703, https://openalex.org/W2525948328, https://openalex.org/W2164510688, https://openalex.org/W2110065479, https://openalex.org/W2913370455, https://openalex.org/W2056636849, https://openalex.org/W2535153710, https://openalex.org/W2742401320, https://openalex.org/W4307819561, https://openalex.org/W2002247284, https://openalex.org/W2898063414, https://openalex.org/W2087921594, https://openalex.org/W2164173275, https://openalex.org/W2290790382, https://openalex.org/W1711578039, https://openalex.org/W2019341789, https://openalex.org/W3102865041, https://openalex.org/W2989340324, https://openalex.org/W2049212652, https://openalex.org/W2164236178, https://openalex.org/W2969263928, https://openalex.org/W3162043068, https://openalex.org/W2036131097, https://openalex.org/W2151380506, https://openalex.org/W2943222680, https://openalex.org/W3201931145, https://openalex.org/W2119123878, https://openalex.org/W4205490067, https://openalex.org/W4210556119, https://openalex.org/W2562701561, https://openalex.org/W2911558244 |
| referenced_works_count | 128 |
| abstract_inverted_index.. | 347 |
| abstract_inverted_index.2 | 83 |
| abstract_inverted_index.a | 114, 175, 193, 219, 259, 266, 342 |
| abstract_inverted_index.(4 | 111 |
| abstract_inverted_index.AD | 333 |
| abstract_inverted_index.As | 56 |
| abstract_inverted_index.At | 192 |
| abstract_inverted_index.ES | 28, 130, 151, 228, 295 |
| abstract_inverted_index.In | 289 |
| abstract_inverted_index.We | 93, 257 |
| abstract_inverted_index.an | 107, 207 |
| abstract_inverted_index.as | 17, 325, 327, 341 |
| abstract_inverted_index.at | 106, 147 |
| abstract_inverted_index.be | 312 |
| abstract_inverted_index.by | 218, 239, 272, 323 |
| abstract_inverted_index.in | 39, 49, 86, 126, 132, 174, 181, 251, 254, 265, 287 |
| abstract_inverted_index.it | 243 |
| abstract_inverted_index.no | 154, 230 |
| abstract_inverted_index.of | 103, 117, 123, 141, 161, 168, 186, 203, 209, 221, 236, 247, 261, 269, 278, 319 |
| abstract_inverted_index.on | 138, 157 |
| abstract_inverted_index.to | 69, 97, 212, 224, 315, 328 |
| abstract_inverted_index.we | 22, 64 |
| abstract_inverted_index.(10 | 197 |
| abstract_inverted_index.Aβ | 213 |
| abstract_inverted_index.ES, | 70, 324 |
| abstract_inverted_index.and | 23, 33, 53, 75, 89, 120, 143, 183, 188, 283, 296, 305 |
| abstract_inverted_index.app | 344 |
| abstract_inverted_index.are | 337 |
| abstract_inverted_index.for | 13, 274, 332 |
| abstract_inverted_index.had | 135, 153 |
| abstract_inverted_index.how | 67 |
| abstract_inverted_index.of, | 32 |
| abstract_inverted_index.our | 291 |
| abstract_inverted_index.the | 11, 30, 57, 72, 79, 100, 139, 158, 169, 199, 233, 245, 275, 279, 316, 320, 329 |
| abstract_inverted_index.to, | 35 |
| abstract_inverted_index.via | 345 |
| abstract_inverted_index.was | 216 |
| abstract_inverted_index.web | 343 |
| abstract_inverted_index.(ES) | 8 |
| abstract_inverted_index.acid | 285 |
| abstract_inverted_index.age, | 149, 241 |
| abstract_inverted_index.ages | 85 |
| abstract_inverted_index.both | 50, 87 |
| abstract_inverted_index.data | 292, 336 |
| abstract_inverted_index.have | 25 |
| abstract_inverted_index.here | 65 |
| abstract_inverted_index.life | 6 |
| abstract_inverted_index.mice | 46, 105, 134, 205 |
| abstract_inverted_index.pre- | 182 |
| abstract_inverted_index.risk | 12, 331 |
| abstract_inverted_index.such | 16 |
| abstract_inverted_index.that | 4, 27, 215, 294 |
| abstract_inverted_index.this | 148, 240 |
| abstract_inverted_index.well | 326 |
| abstract_inverted_index.were | 62 |
| abstract_inverted_index.(AD). | 20 |
| abstract_inverted_index.(Aβ) | 37 |
| abstract_inverted_index.These | 309, 335 |
| abstract_inverted_index.actin | 127 |
| abstract_inverted_index.alpha | 276 |
| abstract_inverted_index.early | 5, 108 |
| abstract_inverted_index.fatty | 284 |
| abstract_inverted_index.fewer | 179 |
| abstract_inverted_index.found | 94 |
| abstract_inverted_index.later | 194 |
| abstract_inverted_index.level | 140 |
| abstract_inverted_index.lipid | 252, 307 |
| abstract_inverted_index.lower | 121 |
| abstract_inverted_index.mice, | 99, 190, 271 |
| abstract_inverted_index.mice. | 92, 164, 256 |
| abstract_inverted_index.might | 311 |
| abstract_inverted_index.share | 298 |
| abstract_inverted_index.shown | 26 |
| abstract_inverted_index.stage | 110, 196 |
| abstract_inverted_index.that, | 95 |
| abstract_inverted_index.these | 60 |
| abstract_inverted_index.using | 71, 171 |
| abstract_inverted_index.across | 82 |
| abstract_inverted_index.animal | 40 |
| abstract_inverted_index.behind | 59 |
| abstract_inverted_index.cohort | 177, 268 |
| abstract_inverted_index.effect | 156 |
| abstract_inverted_index.higher | 115 |
| abstract_inverted_index.levels | 122 |
| abstract_inverted_index.longer | 231 |
| abstract_inverted_index.model, | 77 |
| abstract_inverted_index.online | 340 |
| abstract_inverted_index.others | 24 |
| abstract_inverted_index.showed | 113, 178 |
| abstract_inverted_index.stress | 7 |
| abstract_inverted_index.APP/PS1 | 45, 90, 104, 163, 187, 204, 255, 321 |
| abstract_inverted_index.affects | 78 |
| abstract_inverted_index.animals | 238 |
| abstract_inverted_index.bedding | 76 |
| abstract_inverted_index.changes | 61 |
| abstract_inverted_index.disease | 19 |
| abstract_inverted_index.display | 47 |
| abstract_inverted_index.effects | 137 |
| abstract_inverted_index.limited | 73 |
| abstract_inverted_index.models. | 41 |
| abstract_inverted_index.months) | 112 |
| abstract_inverted_index.nesting | 74 |
| abstract_inverted_index.protein | 263, 282 |
| abstract_inverted_index.related | 211, 223 |
| abstract_inverted_index.similar | 136 |
| abstract_inverted_index.subunit | 277 |
| abstract_inverted_index.suggest | 293 |
| abstract_inverted_index.synapse | 170 |
| abstract_inverted_index.whereas | 150, 242 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.affected | 232, 244 |
| abstract_inverted_index.analysis | 167 |
| abstract_inverted_index.compared | 96 |
| abstract_inverted_index.deficits | 48 |
| abstract_inverted_index.electron | 172 |
| abstract_inverted_index.evidence | 2 |
| abstract_inverted_index.exposure | 9, 68, 131, 152, 229 |
| abstract_inverted_index.involved | 125, 250 |
| abstract_inverted_index.months), | 198 |
| abstract_inverted_index.pathways | 300, 310 |
| abstract_inverted_index.proteins | 119, 124, 146, 210, 222, 249 |
| abstract_inverted_index.proteome | 81, 160, 202, 235 |
| abstract_inverted_index.publicly | 338 |
| abstract_inverted_index.receptor | 226 |
| abstract_inverted_index.response | 34 |
| abstract_inverted_index.revealed | 206 |
| abstract_inverted_index.separate | 267 |
| abstract_inverted_index.synaptic | 54, 80, 201, 234, 302 |
| abstract_inverted_index.synthase | 286 |
| abstract_inverted_index.unclear, | 63 |
| abstract_inverted_index.wildtype | 88, 98, 133, 237 |
| abstract_inverted_index.Moreover, | 42 |
| abstract_inverted_index.abundance | 116 |
| abstract_inverted_index.astrocyte | 262 |
| abstract_inverted_index.cognitive | 51 |
| abstract_inverted_index.confirmed | 258 |
| abstract_inverted_index.different | 84 |
| abstract_inverted_index.diseases, | 15 |
| abstract_inverted_index.dynamics. | 128 |
| abstract_inverted_index.follow-up | 176 |
| abstract_inverted_index.function. | 55 |
| abstract_inverted_index.increases | 10 |
| abstract_inverted_index.indicates | 3 |
| abstract_inverted_index.involving | 301 |
| abstract_inverted_index.long-term | 317 |
| abstract_inverted_index.pathology | 38 |
| abstract_inverted_index.phenotype | 322 |
| abstract_inverted_index.ES-exposed | 43, 189 |
| abstract_inverted_index.accessible | 339 |
| abstract_inverted_index.additional | 155 |
| abstract_inverted_index.aggravates | 29 |
| abstract_inverted_index.astrocytic | 248, 306 |
| abstract_inverted_index.expression | 246, 264 |
| abstract_inverted_index.later-life | 14 |
| abstract_inverted_index.mechanisms | 58 |
| abstract_inverted_index.metabolism | 253 |
| abstract_inverted_index.microscopy | 173 |
| abstract_inverted_index.pathogenic | 299 |
| abstract_inverted_index.transgenic | 44, 91 |
| abstract_inverted_index.underlying | 313 |
| abstract_inverted_index.accompanied | 217 |
| abstract_inverted_index.aggravation | 318 |
| abstract_inverted_index.amyloidosis | 297 |
| abstract_inverted_index.astrocytes. | 288 |
| abstract_inverted_index.conclusion, | 290 |
| abstract_inverted_index.development | 31 |
| abstract_inverted_index.dysfunction | 304 |
| abstract_inverted_index.early-stage | 162 |
| abstract_inverted_index.flexibility | 52 |
| abstract_inverted_index.hippocampal | 101, 200 |
| abstract_inverted_index.metabolism. | 308 |
| abstract_inverted_index.processing, | 214 |
| abstract_inverted_index.12-month-old | 270 |
| abstract_inverted_index.Accordingly, | 21, 165 |
| abstract_inverted_index.amyloid-beta | 36 |
| abstract_inverted_index.compartments | 185 |
| abstract_inverted_index.contributors | 314 |
| abstract_inverted_index.endocytosis. | 227 |
| abstract_inverted_index.investigated | 66 |
| abstract_inverted_index.mitochondria | 180 |
| abstract_inverted_index.pathological | 109, 195 |
| abstract_inverted_index.postsynaptic | 225 |
| abstract_inverted_index.progression. | 334 |
| abstract_inverted_index.synaptosomal | 145, 159 |
| abstract_inverted_index.synaptosomes | 102 |
| abstract_inverted_index.upregulation | 208 |
| abstract_inverted_index.Alzheimer’s | 18 |
| abstract_inverted_index.ES-associated | 330 |
| abstract_inverted_index.actin-related | 144 |
| abstract_inverted_index.dysregulation | 260 |
| abstract_inverted_index.mitochondrial | 118, 142, 280, 303 |
| abstract_inverted_index.post-synaptic | 184 |
| abstract_inverted_index.respectively. | 191 |
| abstract_inverted_index.trifunctional | 281 |
| abstract_inverted_index.Interestingly, | 129 |
| abstract_inverted_index.downregulation | 220 |
| abstract_inverted_index.immunostaining | 273 |
| abstract_inverted_index.Epidemiological | 1 |
| abstract_inverted_index.ultrastructural | 166 |
| abstract_inverted_index.https://amsterdamstudygroup.shinyapps.io/ES_Synaptosome_Proteomics_Visualizer/ | 346 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5076907548 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 13 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.7900000214576721 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.08778551 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |