Effective Molecular Dynamics from Neural-Network Based Structure Prediction Models Article Swipe
 YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.10.17.512476
YOU?
·
· 2022
· Open Access
·
· DOI: https://doi.org/10.1101/2022.10.17.512476
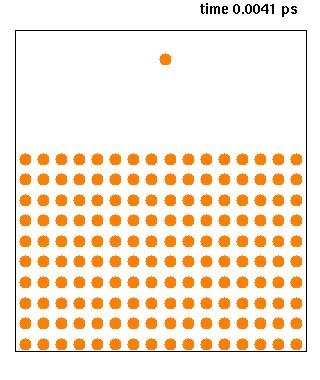
Recent breakthroughs in neural-network based structure prediction methods, such as AlphaFold2 and RoseTTAFold, have dramatically improved the quality of computational protein structure prediction. These models also provide statistical confidence scores that can estimate uncertainties in the predicted structures, but it remains unclear to what extent these scores are related to the intrinsic conformational dynamics of proteins. Here we compare AlphaFold2 prediction scores with >60 μs of explicit molecular dynamics simulations of 28 one- and two-domain proteins with varying degree of flexibility. We demonstrate a strong correlation between the statistical prediction scores and the explicit motion derived from extensive atomistic molecular dynamics simulations, and further derive an elastic network model based on the statistical scores of AlphFold2 (AF-ENM), which we benchmark in combination with coarse-grained molecular dynamics simulations. We show that our AF-ENM method reproduces the global protein dynamics with improved accuracy, providing a powerful way to derive effective molecular dynamics using neural-network based structure prediction models.
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2022.10.17.512476
- https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdf
- OA Status
- green
- Cited By
- 3
- References
- 63
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4306742371
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4306742371Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2022.10.17.512476Digital Object Identifier
- Title
-
Effective Molecular Dynamics from Neural-Network Based Structure Prediction ModelsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-10-18Full publication date if available
- Authors
-
Alexander Jussupow, Ville R. I. KailaList of authors in order
- Landing page
-
https://doi.org/10.1101/2022.10.17.512476Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdfDirect OA link when available
- Concepts
-
Benchmark (surveying), Molecular dynamics, Computer science, Flexibility (engineering), Artificial neural network, Dynamics (music), Protein structure prediction, Statistical physics, Artificial intelligence, Machine learning, Protein structure, Mathematics, Physics, Computational chemistry, Statistics, Chemistry, Geodesy, Nuclear magnetic resonance, Geography, AcousticsTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
3Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1, 2023: 2Per-year citation counts (last 5 years)
- References (count)
-
63Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4306742371 |
|---|---|
| doi | https://doi.org/10.1101/2022.10.17.512476 |
| ids.doi | https://doi.org/10.1101/2022.10.17.512476 |
| ids.openalex | https://openalex.org/W4306742371 |
| fwci | 0.36920747 |
| type | preprint |
| title | Effective Molecular Dynamics from Neural-Network Based Structure Prediction Models |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10044 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Protein Structure and Dynamics |
| topics[1].id | https://openalex.org/T11162 |
| topics[1].field.id | https://openalex.org/fields/25 |
| topics[1].field.display_name | Materials Science |
| topics[1].score | 0.9986000061035156 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2505 |
| topics[1].subfield.display_name | Materials Chemistry |
| topics[1].display_name | Enzyme Structure and Function |
| topics[2].id | https://openalex.org/T10521 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9804999828338623 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA and protein synthesis mechanisms |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C185798385 |
| concepts[0].level | 2 |
| concepts[0].score | 0.6583248376846313 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1161707 |
| concepts[0].display_name | Benchmark (surveying) |
| concepts[1].id | https://openalex.org/C59593255 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6321894526481628 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q901663 |
| concepts[1].display_name | Molecular dynamics |
| concepts[2].id | https://openalex.org/C41008148 |
| concepts[2].level | 0 |
| concepts[2].score | 0.603117823600769 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[2].display_name | Computer science |
| concepts[3].id | https://openalex.org/C2780598303 |
| concepts[3].level | 2 |
| concepts[3].score | 0.6023147106170654 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q65921492 |
| concepts[3].display_name | Flexibility (engineering) |
| concepts[4].id | https://openalex.org/C50644808 |
| concepts[4].level | 2 |
| concepts[4].score | 0.5810354351997375 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q192776 |
| concepts[4].display_name | Artificial neural network |
| concepts[5].id | https://openalex.org/C145912823 |
| concepts[5].level | 2 |
| concepts[5].score | 0.5162491798400879 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q113558 |
| concepts[5].display_name | Dynamics (music) |
| concepts[6].id | https://openalex.org/C18051474 |
| concepts[6].level | 3 |
| concepts[6].score | 0.4806734025478363 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q899656 |
| concepts[6].display_name | Protein structure prediction |
| concepts[7].id | https://openalex.org/C121864883 |
| concepts[7].level | 1 |
| concepts[7].score | 0.3922395408153534 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q677916 |
| concepts[7].display_name | Statistical physics |
| concepts[8].id | https://openalex.org/C154945302 |
| concepts[8].level | 1 |
| concepts[8].score | 0.38550230860710144 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[8].display_name | Artificial intelligence |
| concepts[9].id | https://openalex.org/C119857082 |
| concepts[9].level | 1 |
| concepts[9].score | 0.3434174358844757 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[9].display_name | Machine learning |
| concepts[10].id | https://openalex.org/C47701112 |
| concepts[10].level | 2 |
| concepts[10].score | 0.23989185690879822 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q735188 |
| concepts[10].display_name | Protein structure |
| concepts[11].id | https://openalex.org/C33923547 |
| concepts[11].level | 0 |
| concepts[11].score | 0.17057761549949646 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[11].display_name | Mathematics |
| concepts[12].id | https://openalex.org/C121332964 |
| concepts[12].level | 0 |
| concepts[12].score | 0.14941123127937317 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q413 |
| concepts[12].display_name | Physics |
| concepts[13].id | https://openalex.org/C147597530 |
| concepts[13].level | 1 |
| concepts[13].score | 0.1077607274055481 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q369472 |
| concepts[13].display_name | Computational chemistry |
| concepts[14].id | https://openalex.org/C105795698 |
| concepts[14].level | 1 |
| concepts[14].score | 0.10369053483009338 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[14].display_name | Statistics |
| concepts[15].id | https://openalex.org/C185592680 |
| concepts[15].level | 0 |
| concepts[15].score | 0.10309478640556335 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[15].display_name | Chemistry |
| concepts[16].id | https://openalex.org/C13280743 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q131089 |
| concepts[16].display_name | Geodesy |
| concepts[17].id | https://openalex.org/C46141821 |
| concepts[17].level | 1 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q209402 |
| concepts[17].display_name | Nuclear magnetic resonance |
| concepts[18].id | https://openalex.org/C205649164 |
| concepts[18].level | 0 |
| concepts[18].score | 0.0 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q1071 |
| concepts[18].display_name | Geography |
| concepts[19].id | https://openalex.org/C24890656 |
| concepts[19].level | 1 |
| concepts[19].score | 0.0 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q82811 |
| concepts[19].display_name | Acoustics |
| keywords[0].id | https://openalex.org/keywords/benchmark |
| keywords[0].score | 0.6583248376846313 |
| keywords[0].display_name | Benchmark (surveying) |
| keywords[1].id | https://openalex.org/keywords/molecular-dynamics |
| keywords[1].score | 0.6321894526481628 |
| keywords[1].display_name | Molecular dynamics |
| keywords[2].id | https://openalex.org/keywords/computer-science |
| keywords[2].score | 0.603117823600769 |
| keywords[2].display_name | Computer science |
| keywords[3].id | https://openalex.org/keywords/flexibility |
| keywords[3].score | 0.6023147106170654 |
| keywords[3].display_name | Flexibility (engineering) |
| keywords[4].id | https://openalex.org/keywords/artificial-neural-network |
| keywords[4].score | 0.5810354351997375 |
| keywords[4].display_name | Artificial neural network |
| keywords[5].id | https://openalex.org/keywords/dynamics |
| keywords[5].score | 0.5162491798400879 |
| keywords[5].display_name | Dynamics (music) |
| keywords[6].id | https://openalex.org/keywords/protein-structure-prediction |
| keywords[6].score | 0.4806734025478363 |
| keywords[6].display_name | Protein structure prediction |
| keywords[7].id | https://openalex.org/keywords/statistical-physics |
| keywords[7].score | 0.3922395408153534 |
| keywords[7].display_name | Statistical physics |
| keywords[8].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[8].score | 0.38550230860710144 |
| keywords[8].display_name | Artificial intelligence |
| keywords[9].id | https://openalex.org/keywords/machine-learning |
| keywords[9].score | 0.3434174358844757 |
| keywords[9].display_name | Machine learning |
| keywords[10].id | https://openalex.org/keywords/protein-structure |
| keywords[10].score | 0.23989185690879822 |
| keywords[10].display_name | Protein structure |
| keywords[11].id | https://openalex.org/keywords/mathematics |
| keywords[11].score | 0.17057761549949646 |
| keywords[11].display_name | Mathematics |
| keywords[12].id | https://openalex.org/keywords/physics |
| keywords[12].score | 0.14941123127937317 |
| keywords[12].display_name | Physics |
| keywords[13].id | https://openalex.org/keywords/computational-chemistry |
| keywords[13].score | 0.1077607274055481 |
| keywords[13].display_name | Computational chemistry |
| keywords[14].id | https://openalex.org/keywords/statistics |
| keywords[14].score | 0.10369053483009338 |
| keywords[14].display_name | Statistics |
| keywords[15].id | https://openalex.org/keywords/chemistry |
| keywords[15].score | 0.10309478640556335 |
| keywords[15].display_name | Chemistry |
| language | en |
| locations[0].id | doi:10.1101/2022.10.17.512476 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | cc-by-nc |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2022.10.17.512476 |
| locations[1].id | pmh:oai:figshare.com:article/22335560 |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S4306400572 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[1].source.host_organization | https://openalex.org/I196829312 |
| locations[1].source.host_organization_name | La Trobe University |
| locations[1].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[1].license | cc-by-nc |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | Text |
| locations[1].license_id | https://openalex.org/licenses/cc-by-nc |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://figshare.com/articles/journal_contribution/Effective_Molecular_Dynamics_from_Neural_Network-Based_Structure_Prediction_Models/22335560 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5045312855 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-7851-2741 |
| authorships[0].author.display_name | Alexander Jussupow |
| authorships[0].countries | SE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I161593684 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Biochemistry and Biophysics, Stockholm University, 069, Stockholm, Sweden |
| authorships[0].institutions[0].id | https://openalex.org/I161593684 |
| authorships[0].institutions[0].ror | https://ror.org/05f0yaq80 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I161593684 |
| authorships[0].institutions[0].country_code | SE |
| authorships[0].institutions[0].display_name | Stockholm University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Alexander Jussupow |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Biochemistry and Biophysics, Stockholm University, 069, Stockholm, Sweden |
| authorships[1].author.id | https://openalex.org/A5063740635 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-4464-6324 |
| authorships[1].author.display_name | Ville R. I. Kaila |
| authorships[1].countries | SE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I161593684 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Biochemistry and Biophysics, Stockholm University, 069, Stockholm, Sweden |
| authorships[1].institutions[0].id | https://openalex.org/I161593684 |
| authorships[1].institutions[0].ror | https://ror.org/05f0yaq80 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I161593684 |
| authorships[1].institutions[0].country_code | SE |
| authorships[1].institutions[0].display_name | Stockholm University |
| authorships[1].author_position | last |
| authorships[1].raw_author_name | Ville R. I. Kaila |
| authorships[1].is_corresponding | True |
| authorships[1].raw_affiliation_strings | Department of Biochemistry and Biophysics, Stockholm University, 069, Stockholm, Sweden |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Effective Molecular Dynamics from Neural-Network Based Structure Prediction Models |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10044 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Protein Structure and Dynamics |
| related_works | https://openalex.org/W2378211422, https://openalex.org/W4321353415, https://openalex.org/W2745001401, https://openalex.org/W2130974462, https://openalex.org/W2028665553, https://openalex.org/W2086519370, https://openalex.org/W4246352526, https://openalex.org/W2121910908, https://openalex.org/W915438175, https://openalex.org/W1773535258 |
| cited_by_count | 3 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| counts_by_year[1].year | 2023 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1101/2022.10.17.512476 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | cc-by-nc |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by-nc |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2022.10.17.512476 |
| primary_location.id | doi:10.1101/2022.10.17.512476 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | cc-by-nc |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2022/10/18/2022.10.17.512476.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by-nc |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2022.10.17.512476 |
| publication_date | 2022-10-18 |
| publication_year | 2022 |
| referenced_works | https://openalex.org/W3177828909, https://openalex.org/W3186179742, https://openalex.org/W3211795435, https://openalex.org/W4288428465, https://openalex.org/W4213255340, https://openalex.org/W2950954328, https://openalex.org/W3202105508, https://openalex.org/W4304731451, https://openalex.org/W2140673705, https://openalex.org/W3183475563, https://openalex.org/W3204490060, https://openalex.org/W3195375135, https://openalex.org/W4224126566, https://openalex.org/W4213208821, https://openalex.org/W4214861623, https://openalex.org/W2161151688, https://openalex.org/W2110483430, https://openalex.org/W3205544459, https://openalex.org/W4229373389, https://openalex.org/W4214847177, https://openalex.org/W2156016724, https://openalex.org/W2891656222, https://openalex.org/W2142532813, https://openalex.org/W2051864856, https://openalex.org/W2069777230, https://openalex.org/W2413334978, https://openalex.org/W2119267663, https://openalex.org/W2314292783, https://openalex.org/W3148042916, https://openalex.org/W2081724315, https://openalex.org/W3169781269, https://openalex.org/W2401568999, https://openalex.org/W2119806687, https://openalex.org/W4205688077, https://openalex.org/W2065947633, https://openalex.org/W2768114066, https://openalex.org/W3020669571, https://openalex.org/W3093420066, https://openalex.org/W4223417529, https://openalex.org/W1970729662, https://openalex.org/W2588157903, https://openalex.org/W4281686845, https://openalex.org/W2800697076, https://openalex.org/W2062058722, https://openalex.org/W2120702846, https://openalex.org/W1998425418, https://openalex.org/W3012254059, https://openalex.org/W3097536747, https://openalex.org/W4221028205, https://openalex.org/W2078646305, https://openalex.org/W2131377322, https://openalex.org/W2320151114, https://openalex.org/W1990235290, https://openalex.org/W2127215376, https://openalex.org/W2292301738, https://openalex.org/W2294798173, https://openalex.org/W1965555277, https://openalex.org/W2001150023, https://openalex.org/W2101017457, https://openalex.org/W1031578623, https://openalex.org/W2057477511, https://openalex.org/W2081693079, https://openalex.org/W3013283700 |
| referenced_works_count | 63 |
| abstract_inverted_index.a | 84, 143 |
| abstract_inverted_index.28 | 72 |
| abstract_inverted_index.We | 82, 128 |
| abstract_inverted_index.an | 106 |
| abstract_inverted_index.as | 10 |
| abstract_inverted_index.in | 3, 35, 121 |
| abstract_inverted_index.it | 40 |
| abstract_inverted_index.of | 19, 55, 66, 71, 80, 115 |
| abstract_inverted_index.on | 111 |
| abstract_inverted_index.to | 43, 50, 146 |
| abstract_inverted_index.we | 58, 119 |
| abstract_inverted_index.and | 12, 74, 92, 103 |
| abstract_inverted_index.are | 48 |
| abstract_inverted_index.but | 39 |
| abstract_inverted_index.can | 32 |
| abstract_inverted_index.our | 131 |
| abstract_inverted_index.the | 17, 36, 51, 88, 93, 112, 135 |
| abstract_inverted_index.way | 145 |
| abstract_inverted_index.μs | 65 |
| abstract_inverted_index.Here | 57 |
| abstract_inverted_index.also | 26 |
| abstract_inverted_index.from | 97 |
| abstract_inverted_index.have | 14 |
| abstract_inverted_index.one- | 73 |
| abstract_inverted_index.show | 129 |
| abstract_inverted_index.such | 9 |
| abstract_inverted_index.that | 31, 130 |
| abstract_inverted_index.what | 44 |
| abstract_inverted_index.with | 63, 77, 123, 139 |
| abstract_inverted_index.These | 24 |
| abstract_inverted_index.based | 5, 110, 153 |
| abstract_inverted_index.model | 109 |
| abstract_inverted_index.these | 46 |
| abstract_inverted_index.using | 151 |
| abstract_inverted_index.which | 118 |
| abstract_inverted_index.>60 | 64 |
| abstract_inverted_index.AF-ENM | 132 |
| abstract_inverted_index.Recent | 1 |
| abstract_inverted_index.degree | 79 |
| abstract_inverted_index.derive | 105, 147 |
| abstract_inverted_index.extent | 45 |
| abstract_inverted_index.global | 136 |
| abstract_inverted_index.method | 133 |
| abstract_inverted_index.models | 25 |
| abstract_inverted_index.motion | 95 |
| abstract_inverted_index.scores | 30, 47, 62, 91, 114 |
| abstract_inverted_index.strong | 85 |
| abstract_inverted_index.between | 87 |
| abstract_inverted_index.compare | 59 |
| abstract_inverted_index.derived | 96 |
| abstract_inverted_index.elastic | 107 |
| abstract_inverted_index.further | 104 |
| abstract_inverted_index.models. | 156 |
| abstract_inverted_index.network | 108 |
| abstract_inverted_index.protein | 21, 137 |
| abstract_inverted_index.provide | 27 |
| abstract_inverted_index.quality | 18 |
| abstract_inverted_index.related | 49 |
| abstract_inverted_index.remains | 41 |
| abstract_inverted_index.unclear | 42 |
| abstract_inverted_index.varying | 78 |
| abstract_inverted_index.ABSTRACT | 0 |
| abstract_inverted_index.dynamics | 54, 69, 101, 126, 138, 150 |
| abstract_inverted_index.estimate | 33 |
| abstract_inverted_index.explicit | 67, 94 |
| abstract_inverted_index.improved | 16, 140 |
| abstract_inverted_index.methods, | 8 |
| abstract_inverted_index.powerful | 144 |
| abstract_inverted_index.proteins | 76 |
| abstract_inverted_index.(AF-ENM), | 117 |
| abstract_inverted_index.AlphFold2 | 116 |
| abstract_inverted_index.accuracy, | 141 |
| abstract_inverted_index.atomistic | 99 |
| abstract_inverted_index.benchmark | 120 |
| abstract_inverted_index.effective | 148 |
| abstract_inverted_index.extensive | 98 |
| abstract_inverted_index.intrinsic | 52 |
| abstract_inverted_index.molecular | 68, 100, 125, 149 |
| abstract_inverted_index.predicted | 37 |
| abstract_inverted_index.proteins. | 56 |
| abstract_inverted_index.providing | 142 |
| abstract_inverted_index.structure | 6, 22, 154 |
| abstract_inverted_index.AlphaFold2 | 11, 60 |
| abstract_inverted_index.confidence | 29 |
| abstract_inverted_index.prediction | 7, 61, 90, 155 |
| abstract_inverted_index.reproduces | 134 |
| abstract_inverted_index.two-domain | 75 |
| abstract_inverted_index.combination | 122 |
| abstract_inverted_index.correlation | 86 |
| abstract_inverted_index.demonstrate | 83 |
| abstract_inverted_index.prediction. | 23 |
| abstract_inverted_index.simulations | 70 |
| abstract_inverted_index.statistical | 28, 89, 113 |
| abstract_inverted_index.structures, | 38 |
| abstract_inverted_index.RoseTTAFold, | 13 |
| abstract_inverted_index.dramatically | 15 |
| abstract_inverted_index.flexibility. | 81 |
| abstract_inverted_index.simulations, | 102 |
| abstract_inverted_index.simulations. | 127 |
| abstract_inverted_index.breakthroughs | 2 |
| abstract_inverted_index.computational | 20 |
| abstract_inverted_index.uncertainties | 34 |
| abstract_inverted_index.coarse-grained | 124 |
| abstract_inverted_index.conformational | 53 |
| abstract_inverted_index.neural-network | 4, 152 |
| cited_by_percentile_year.max | 96 |
| cited_by_percentile_year.min | 91 |
| corresponding_author_ids | https://openalex.org/A5063740635 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 2 |
| corresponding_institution_ids | https://openalex.org/I161593684 |
| citation_normalized_percentile.value | 0.55299647 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |