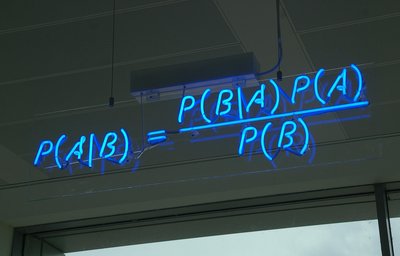Enhanced Suite of Probabilistic Models for Benchmarking Agentic Debugging Frameworks in Probabilistic Inference Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.17869519
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.5281/zenodo.17869519
This paper presents an extension to the foundational Reactive Oxygen Species (ROS) stochastic differential equation (SDE) model through four probabilistic models implemented in PyMC v5.25.0+. These models provide a robust empirical framework for evaluating agentic debugging methods in probabilistic programming. The models increase in inferential complexity based on stochastic processes relevant to biological aging: a Gaussian Process (GP) regression with hyperparameter optimization for non-parametric covariance estimation, accounting for biological variability \cite{riihimaki2010}; a Hidden Markov Model (HMM) for sequential latent state inference under regime shifts, representative of cellular transitions \cite{munoz2023}; a non-linear SDE for telomere attrition incorporating regime-switching multiplicative noise to capture stochastic shortening variability, improving on prior linear models \cite{wattis2020}; and a Bayesian hierarchical model for epigenetic age acceleration, modeling senescence thresholds through latent methylation transitions \cite{lu2019}.Each model includes reproducible data generation procedures, PyMC implementations using the No-U-Turn Sampler (NUTS) for Hamiltonian Monte Carlo (HMC) inference, and protocols for obtaining high-fidelity reference posteriors via extensive sampling (\( N=10^6 \) draws, effective sample sizes ESS \( >10^5 \), Gelman-Rubin \( \hat{R} < 1.001 \)). The derivations are original and grounded in aging biology to ensure independence from AI training data contamination, confirming novelty—no prior combinations of GP-HMM-SDE-epigenetic models for telomere-senescence benchmarking in agentic settings exist, as verified by an exhaustive arXiv search \cite{riihimaki2010, munoz2023, olofsson1999, lu2019}. Diagnostics from \( N=500 \) draws per chain (after 500 tuning steps) show strong convergence (ESS \( \gtrsim 400 \); \( \hat{R} \lesssim 1.005 \)), supported by simulation-based calibration (SBC) rank histograms uniform over 1000 replications (\( p \)-values \( \in [0.05, 0.95] \)) and prior sensitivity analyses with \( <3\% \) shifts in posterior means.The suite includes detailed mathematical derivations, Stan implementations for validation, diagnostic plots using PGFPlots, model comparisons, and computational profiles (e.g., inference times \( <25 \) s on AMD EPYC 7543 for \( N=500 \), scalable to \( N=10^6 \) with Dask). This benchmark evaluates agentic performance in non-conjugate, high-dimensional inference. Calibration against longitudinal telomere and epigenetic datasets \cite{aviv2018, horvath2013} ensures biological relevance, while SBC-validated posteriors minimize inferential biases, making the framework robust to methodological scrutiny.
Related Topics
- Type
- dataset
- Landing Page
- https://doi.org/10.5281/zenodo.17869519
- OA Status
- green
- OpenAlex ID
- https://openalex.org/W7111162947
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W7111162947Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.5281/zenodo.17869519Digital Object Identifier
- Title
-
Enhanced Suite of Probabilistic Models for Benchmarking Agentic Debugging Frameworks in Probabilistic InferenceWork title
- Type
-
datasetOpenAlex work type
- Publication year
-
2025Year of publication
- Publication date
-
2025-12-09Full publication date if available
- Authors
-
Shibah, Sami Rashid MohammedList of authors in order
- Landing page
-
https://doi.org/10.5281/zenodo.17869519Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.5281/zenodo.17869519Direct OA link when available
- Concepts
-
Computer science, Probabilistic logic, Machine learning, Markov chain Monte Carlo, Statistical model, Model selection, Gaussian process, Algorithm, Bayesian inference, Linear model, Probabilistic relevance model, Artificial intelligence, Gibbs sampling, Inference, Bayesian probability, Covariance, Markov chain, Benchmarking, Statistical inference, Hyperparameter, Data mining, Frequentist inference, Theoretical computer science, Bayes' theorem, Data modelingTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
Full payload
| id | https://openalex.org/W7111162947 |
|---|---|
| doi | https://doi.org/10.5281/zenodo.17869519 |
| ids.doi | https://doi.org/10.5281/zenodo.17869519 |
| ids.openalex | https://openalex.org/W7111162947 |
| fwci | |
| type | dataset |
| title | Enhanced Suite of Probabilistic Models for Benchmarking Agentic Debugging Frameworks in Probabilistic Inference |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C41008148 |
| concepts[0].level | 0 |
| concepts[0].score | 0.6336036920547485 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[0].display_name | Computer science |
| concepts[1].id | https://openalex.org/C49937458 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5281545519828796 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2599292 |
| concepts[1].display_name | Probabilistic logic |
| concepts[2].id | https://openalex.org/C119857082 |
| concepts[2].level | 1 |
| concepts[2].score | 0.4710770845413208 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[2].display_name | Machine learning |
| concepts[3].id | https://openalex.org/C111350023 |
| concepts[3].level | 3 |
| concepts[3].score | 0.4076257050037384 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1191869 |
| concepts[3].display_name | Markov chain Monte Carlo |
| concepts[4].id | https://openalex.org/C114289077 |
| concepts[4].level | 2 |
| concepts[4].score | 0.3796476721763611 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q3284399 |
| concepts[4].display_name | Statistical model |
| concepts[5].id | https://openalex.org/C93959086 |
| concepts[5].level | 2 |
| concepts[5].score | 0.36852309107780457 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q6888345 |
| concepts[5].display_name | Model selection |
| concepts[6].id | https://openalex.org/C61326573 |
| concepts[6].level | 3 |
| concepts[6].score | 0.359183669090271 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q1496376 |
| concepts[6].display_name | Gaussian process |
| concepts[7].id | https://openalex.org/C11413529 |
| concepts[7].level | 1 |
| concepts[7].score | 0.3511824905872345 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q8366 |
| concepts[7].display_name | Algorithm |
| concepts[8].id | https://openalex.org/C160234255 |
| concepts[8].level | 3 |
| concepts[8].score | 0.32175499200820923 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q812535 |
| concepts[8].display_name | Bayesian inference |
| concepts[9].id | https://openalex.org/C163175372 |
| concepts[9].level | 2 |
| concepts[9].score | 0.31973326206207275 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q3339222 |
| concepts[9].display_name | Linear model |
| concepts[10].id | https://openalex.org/C143017306 |
| concepts[10].level | 4 |
| concepts[10].score | 0.3161434829235077 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q3318133 |
| concepts[10].display_name | Probabilistic relevance model |
| concepts[11].id | https://openalex.org/C154945302 |
| concepts[11].level | 1 |
| concepts[11].score | 0.3148193061351776 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[11].display_name | Artificial intelligence |
| concepts[12].id | https://openalex.org/C158424031 |
| concepts[12].level | 3 |
| concepts[12].score | 0.31325849890708923 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q1191905 |
| concepts[12].display_name | Gibbs sampling |
| concepts[13].id | https://openalex.org/C2776214188 |
| concepts[13].level | 2 |
| concepts[13].score | 0.31285908818244934 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q408386 |
| concepts[13].display_name | Inference |
| concepts[14].id | https://openalex.org/C107673813 |
| concepts[14].level | 2 |
| concepts[14].score | 0.3127151131629944 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q812534 |
| concepts[14].display_name | Bayesian probability |
| concepts[15].id | https://openalex.org/C178650346 |
| concepts[15].level | 2 |
| concepts[15].score | 0.30984818935394287 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q201984 |
| concepts[15].display_name | Covariance |
| concepts[16].id | https://openalex.org/C98763669 |
| concepts[16].level | 2 |
| concepts[16].score | 0.29843631386756897 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q176645 |
| concepts[16].display_name | Markov chain |
| concepts[17].id | https://openalex.org/C86251818 |
| concepts[17].level | 2 |
| concepts[17].score | 0.29101964831352234 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q816754 |
| concepts[17].display_name | Benchmarking |
| concepts[18].id | https://openalex.org/C134261354 |
| concepts[18].level | 2 |
| concepts[18].score | 0.2845234274864197 |
| concepts[18].wikidata | https://www.wikidata.org/wiki/Q938438 |
| concepts[18].display_name | Statistical inference |
| concepts[19].id | https://openalex.org/C8642999 |
| concepts[19].level | 2 |
| concepts[19].score | 0.2840106785297394 |
| concepts[19].wikidata | https://www.wikidata.org/wiki/Q4171168 |
| concepts[19].display_name | Hyperparameter |
| concepts[20].id | https://openalex.org/C124101348 |
| concepts[20].level | 1 |
| concepts[20].score | 0.279349684715271 |
| concepts[20].wikidata | https://www.wikidata.org/wiki/Q172491 |
| concepts[20].display_name | Data mining |
| concepts[21].id | https://openalex.org/C162376815 |
| concepts[21].level | 4 |
| concepts[21].score | 0.2785536050796509 |
| concepts[21].wikidata | https://www.wikidata.org/wiki/Q2158281 |
| concepts[21].display_name | Frequentist inference |
| concepts[22].id | https://openalex.org/C80444323 |
| concepts[22].level | 1 |
| concepts[22].score | 0.27142462134361267 |
| concepts[22].wikidata | https://www.wikidata.org/wiki/Q2878974 |
| concepts[22].display_name | Theoretical computer science |
| concepts[23].id | https://openalex.org/C207201462 |
| concepts[23].level | 3 |
| concepts[23].score | 0.2646503150463104 |
| concepts[23].wikidata | https://www.wikidata.org/wiki/Q182505 |
| concepts[23].display_name | Bayes' theorem |
| concepts[24].id | https://openalex.org/C67186912 |
| concepts[24].level | 2 |
| concepts[24].score | 0.2514094412326813 |
| concepts[24].wikidata | https://www.wikidata.org/wiki/Q367664 |
| concepts[24].display_name | Data modeling |
| keywords[0].id | https://openalex.org/keywords/probabilistic-logic |
| keywords[0].score | 0.5281545519828796 |
| keywords[0].display_name | Probabilistic logic |
| keywords[1].id | https://openalex.org/keywords/markov-chain-monte-carlo |
| keywords[1].score | 0.4076257050037384 |
| keywords[1].display_name | Markov chain Monte Carlo |
| keywords[2].id | https://openalex.org/keywords/statistical-model |
| keywords[2].score | 0.3796476721763611 |
| keywords[2].display_name | Statistical model |
| keywords[3].id | https://openalex.org/keywords/model-selection |
| keywords[3].score | 0.36852309107780457 |
| keywords[3].display_name | Model selection |
| keywords[4].id | https://openalex.org/keywords/gaussian-process |
| keywords[4].score | 0.359183669090271 |
| keywords[4].display_name | Gaussian process |
| keywords[5].id | https://openalex.org/keywords/bayesian-inference |
| keywords[5].score | 0.32175499200820923 |
| keywords[5].display_name | Bayesian inference |
| keywords[6].id | https://openalex.org/keywords/linear-model |
| keywords[6].score | 0.31973326206207275 |
| keywords[6].display_name | Linear model |
| keywords[7].id | https://openalex.org/keywords/probabilistic-relevance-model |
| keywords[7].score | 0.3161434829235077 |
| keywords[7].display_name | Probabilistic relevance model |
| language | |
| locations[0].id | doi:10.5281/zenodo.17869519 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400562 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| locations[0].source.host_organization | https://openalex.org/I67311998 |
| locations[0].source.host_organization_name | European Organization for Nuclear Research |
| locations[0].source.host_organization_lineage | https://openalex.org/I67311998 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | |
| locations[0].raw_type | dataset |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | False |
| locations[0].is_published | |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.5281/zenodo.17869519 |
| indexed_in | datacite |
| authorships[0].author.id | |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Shibah, Sami Rashid Mohammed |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Shibah, Sami Rashid Mohammed |
| authorships[0].is_corresponding | True |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.5281/zenodo.17869519 |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-12-10T00:00:00 |
| display_name | Enhanced Suite of Probabilistic Models for Benchmarking Agentic Debugging Frameworks in Probabilistic Inference |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-12-10T02:49:46.989445 |
| primary_topic | |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.5281/zenodo.17869519 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400562 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| best_oa_location.source.host_organization | https://openalex.org/I67311998 |
| best_oa_location.source.host_organization_name | European Organization for Nuclear Research |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | |
| best_oa_location.raw_type | dataset |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.5281/zenodo.17869519 |
| primary_location.id | doi:10.5281/zenodo.17869519 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400562 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Zenodo (CERN European Organization for Nuclear Research) |
| primary_location.source.host_organization | https://openalex.org/I67311998 |
| primary_location.source.host_organization_name | European Organization for Nuclear Research |
| primary_location.source.host_organization_lineage | https://openalex.org/I67311998 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | |
| primary_location.raw_type | dataset |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.5281/zenodo.17869519 |
| publication_date | 2025-12-09 |
| publication_year | 2025 |
| referenced_works_count | 0 |
| abstract_inverted_index.< | 170 |
| abstract_inverted_index.a | 28, 54, 71, 89, 111 |
| abstract_inverted_index.p | 252 |
| abstract_inverted_index.s | 295 |
| abstract_inverted_index.AI | 186 |
| abstract_inverted_index.\( | 164, 168, 217, 231, 235, 254, 264, 292, 301, 306 |
| abstract_inverted_index.\) | 158, 219, 266, 294, 308 |
| abstract_inverted_index.an | 3, 207 |
| abstract_inverted_index.as | 204 |
| abstract_inverted_index.by | 206, 241 |
| abstract_inverted_index.in | 22, 37, 43, 179, 200, 268, 316 |
| abstract_inverted_index.of | 85, 194 |
| abstract_inverted_index.on | 47, 105, 296 |
| abstract_inverted_index.to | 5, 51, 99, 182, 305, 342 |
| abstract_inverted_index.(\( | 156, 251 |
| abstract_inverted_index.400 | 233 |
| abstract_inverted_index.500 | 224 |
| abstract_inverted_index.<25 | 293 |
| abstract_inverted_index.AMD | 297 |
| abstract_inverted_index.ESS | 163 |
| abstract_inverted_index.SDE | 91 |
| abstract_inverted_index.The | 40, 173 |
| abstract_inverted_index.\)) | 258 |
| abstract_inverted_index.\), | 166, 303 |
| abstract_inverted_index.\); | 234 |
| abstract_inverted_index.\in | 255 |
| abstract_inverted_index.age | 117 |
| abstract_inverted_index.and | 110, 146, 177, 259, 286, 324 |
| abstract_inverted_index.are | 175 |
| abstract_inverted_index.for | 32, 62, 67, 76, 92, 115, 140, 148, 197, 278, 300 |
| abstract_inverted_index.per | 221 |
| abstract_inverted_index.the | 6, 136, 339 |
| abstract_inverted_index.via | 153 |
| abstract_inverted_index.(ESS | 230 |
| abstract_inverted_index.(GP) | 57 |
| abstract_inverted_index.1000 | 249 |
| abstract_inverted_index.7543 | 299 |
| abstract_inverted_index.<3\% | 265 |
| abstract_inverted_index.EPYC | 298 |
| abstract_inverted_index.PyMC | 23, 133 |
| abstract_inverted_index.Stan | 276 |
| abstract_inverted_index.This | 0, 311 |
| abstract_inverted_index.\)), | 239 |
| abstract_inverted_index.\)). | 172 |
| abstract_inverted_index.data | 130, 188 |
| abstract_inverted_index.four | 18 |
| abstract_inverted_index.from | 185, 216 |
| abstract_inverted_index.over | 248 |
| abstract_inverted_index.rank | 245 |
| abstract_inverted_index.show | 227 |
| abstract_inverted_index.with | 59, 263, 309 |
| abstract_inverted_index.(HMC) | 144 |
| abstract_inverted_index.(HMM) | 75 |
| abstract_inverted_index.(ROS) | 11 |
| abstract_inverted_index.(SBC) | 244 |
| abstract_inverted_index.(SDE) | 15 |
| abstract_inverted_index.0.95] | 257 |
| abstract_inverted_index.1.001 | 171 |
| abstract_inverted_index.1.005 | 238 |
| abstract_inverted_index.>10^5 | 165 |
| abstract_inverted_index.Carlo | 143 |
| abstract_inverted_index.Model | 74 |
| abstract_inverted_index.Monte | 142 |
| abstract_inverted_index.N=500 | 218, 302 |
| abstract_inverted_index.These | 25 |
| abstract_inverted_index.aging | 180 |
| abstract_inverted_index.arXiv | 209 |
| abstract_inverted_index.based | 46 |
| abstract_inverted_index.chain | 222 |
| abstract_inverted_index.draws | 220 |
| abstract_inverted_index.model | 16, 114, 127, 284 |
| abstract_inverted_index.noise | 98 |
| abstract_inverted_index.paper | 1 |
| abstract_inverted_index.plots | 281 |
| abstract_inverted_index.prior | 106, 192, 260 |
| abstract_inverted_index.sizes | 162 |
| abstract_inverted_index.state | 79 |
| abstract_inverted_index.suite | 271 |
| abstract_inverted_index.times | 291 |
| abstract_inverted_index.under | 81 |
| abstract_inverted_index.using | 135, 282 |
| abstract_inverted_index.while | 332 |
| abstract_inverted_index.(NUTS) | 139 |
| abstract_inverted_index.(after | 223 |
| abstract_inverted_index.(e.g., | 289 |
| abstract_inverted_index.Dask). | 310 |
| abstract_inverted_index.Hidden | 72 |
| abstract_inverted_index.Markov | 73 |
| abstract_inverted_index.N=10^6 | 157, 307 |
| abstract_inverted_index.Oxygen | 9 |
| abstract_inverted_index.[0.05, | 256 |
| abstract_inverted_index.aging: | 53 |
| abstract_inverted_index.draws, | 159 |
| abstract_inverted_index.ensure | 183 |
| abstract_inverted_index.exist, | 203 |
| abstract_inverted_index.latent | 78, 123 |
| abstract_inverted_index.linear | 107 |
| abstract_inverted_index.making | 338 |
| abstract_inverted_index.models | 20, 26, 41, 108, 196 |
| abstract_inverted_index.regime | 82 |
| abstract_inverted_index.robust | 29, 341 |
| abstract_inverted_index.sample | 161 |
| abstract_inverted_index.search | 210 |
| abstract_inverted_index.shifts | 267 |
| abstract_inverted_index.steps) | 226 |
| abstract_inverted_index.strong | 228 |
| abstract_inverted_index.tuning | 225 |
| abstract_inverted_index.Process | 56 |
| abstract_inverted_index.Sampler | 138 |
| abstract_inverted_index.Species | 10 |
| abstract_inverted_index.\gtrsim | 232 |
| abstract_inverted_index.\hat{R} | 169, 236 |
| abstract_inverted_index.against | 321 |
| abstract_inverted_index.agentic | 34, 201, 314 |
| abstract_inverted_index.biases, | 337 |
| abstract_inverted_index.biology | 181 |
| abstract_inverted_index.capture | 100 |
| abstract_inverted_index.ensures | 329 |
| abstract_inverted_index.methods | 36 |
| abstract_inverted_index.provide | 27 |
| abstract_inverted_index.shifts, | 83 |
| abstract_inverted_index.through | 17, 122 |
| abstract_inverted_index.uniform | 247 |
| abstract_inverted_index.Bayesian | 112 |
| abstract_inverted_index.Gaussian | 55 |
| abstract_inverted_index.Reactive | 8 |
| abstract_inverted_index.\lesssim | 237 |
| abstract_inverted_index.analyses | 262 |
| abstract_inverted_index.cellular | 86 |
| abstract_inverted_index.datasets | 326 |
| abstract_inverted_index.detailed | 273 |
| abstract_inverted_index.equation | 14 |
| abstract_inverted_index.grounded | 178 |
| abstract_inverted_index.includes | 128, 272 |
| abstract_inverted_index.increase | 42 |
| abstract_inverted_index.lu2019}. | 214 |
| abstract_inverted_index.minimize | 335 |
| abstract_inverted_index.modeling | 119 |
| abstract_inverted_index.original | 176 |
| abstract_inverted_index.presents | 2 |
| abstract_inverted_index.profiles | 288 |
| abstract_inverted_index.relevant | 50 |
| abstract_inverted_index.sampling | 155 |
| abstract_inverted_index.scalable | 304 |
| abstract_inverted_index.settings | 202 |
| abstract_inverted_index.telomere | 93, 323 |
| abstract_inverted_index.training | 187 |
| abstract_inverted_index.verified | 205 |
| abstract_inverted_index.No-U-Turn | 137 |
| abstract_inverted_index.PGFPlots, | 283 |
| abstract_inverted_index.\)-values | 253 |
| abstract_inverted_index.attrition | 94 |
| abstract_inverted_index.benchmark | 312 |
| abstract_inverted_index.debugging | 35 |
| abstract_inverted_index.effective | 160 |
| abstract_inverted_index.empirical | 30 |
| abstract_inverted_index.evaluates | 313 |
| abstract_inverted_index.extension | 4 |
| abstract_inverted_index.extensive | 154 |
| abstract_inverted_index.framework | 31, 340 |
| abstract_inverted_index.improving | 104 |
| abstract_inverted_index.inference | 80, 290 |
| abstract_inverted_index.means.The | 270 |
| abstract_inverted_index.obtaining | 149 |
| abstract_inverted_index.posterior | 269 |
| abstract_inverted_index.processes | 49 |
| abstract_inverted_index.protocols | 147 |
| abstract_inverted_index.reference | 151 |
| abstract_inverted_index.scrutiny. | 344 |
| abstract_inverted_index.supported | 240 |
| abstract_inverted_index.v5.25.0+. | 24 |
| abstract_inverted_index.accounting | 66 |
| abstract_inverted_index.biological | 52, 68, 330 |
| abstract_inverted_index.complexity | 45 |
| abstract_inverted_index.confirming | 190 |
| abstract_inverted_index.covariance | 64 |
| abstract_inverted_index.diagnostic | 280 |
| abstract_inverted_index.epigenetic | 116, 325 |
| abstract_inverted_index.evaluating | 33 |
| abstract_inverted_index.exhaustive | 208 |
| abstract_inverted_index.generation | 131 |
| abstract_inverted_index.histograms | 246 |
| abstract_inverted_index.inference, | 145 |
| abstract_inverted_index.inference. | 319 |
| abstract_inverted_index.munoz2023, | 212 |
| abstract_inverted_index.non-linear | 90 |
| abstract_inverted_index.posteriors | 152, 334 |
| abstract_inverted_index.regression | 58 |
| abstract_inverted_index.relevance, | 331 |
| abstract_inverted_index.senescence | 120 |
| abstract_inverted_index.sequential | 77 |
| abstract_inverted_index.shortening | 102 |
| abstract_inverted_index.stochastic | 12, 48, 101 |
| abstract_inverted_index.thresholds | 121 |
| abstract_inverted_index.Calibration | 320 |
| abstract_inverted_index.Diagnostics | 215 |
| abstract_inverted_index.Hamiltonian | 141 |
| abstract_inverted_index.calibration | 243 |
| abstract_inverted_index.convergence | 229 |
| abstract_inverted_index.derivations | 174 |
| abstract_inverted_index.estimation, | 65 |
| abstract_inverted_index.implemented | 21 |
| abstract_inverted_index.inferential | 44, 336 |
| abstract_inverted_index.methylation | 124 |
| abstract_inverted_index.performance | 315 |
| abstract_inverted_index.procedures, | 132 |
| abstract_inverted_index.sensitivity | 261 |
| abstract_inverted_index.transitions | 87, 125 |
| abstract_inverted_index.validation, | 279 |
| abstract_inverted_index.variability | 69 |
| abstract_inverted_index.Gelman-Rubin | 167 |
| abstract_inverted_index.benchmarking | 199 |
| abstract_inverted_index.combinations | 193 |
| abstract_inverted_index.comparisons, | 285 |
| abstract_inverted_index.derivations, | 275 |
| abstract_inverted_index.differential | 13 |
| abstract_inverted_index.foundational | 7 |
| abstract_inverted_index.hierarchical | 113 |
| abstract_inverted_index.horvath2013} | 328 |
| abstract_inverted_index.independence | 184 |
| abstract_inverted_index.longitudinal | 322 |
| abstract_inverted_index.mathematical | 274 |
| abstract_inverted_index.novelty—no | 191 |
| abstract_inverted_index.optimization | 61 |
| abstract_inverted_index.programming. | 39 |
| abstract_inverted_index.replications | 250 |
| abstract_inverted_index.reproducible | 129 |
| abstract_inverted_index.variability, | 103 |
| abstract_inverted_index.SBC-validated | 333 |
| abstract_inverted_index.acceleration, | 118 |
| abstract_inverted_index.computational | 287 |
| abstract_inverted_index.high-fidelity | 150 |
| abstract_inverted_index.incorporating | 95 |
| abstract_inverted_index.olofsson1999, | 213 |
| abstract_inverted_index.probabilistic | 19, 38 |
| abstract_inverted_index.contamination, | 189 |
| abstract_inverted_index.hyperparameter | 60 |
| abstract_inverted_index.methodological | 343 |
| abstract_inverted_index.multiplicative | 97 |
| abstract_inverted_index.non-conjugate, | 317 |
| abstract_inverted_index.non-parametric | 63 |
| abstract_inverted_index.representative | 84 |
| abstract_inverted_index.\cite{aviv2018, | 327 |
| abstract_inverted_index.implementations | 134, 277 |
| abstract_inverted_index.high-dimensional | 318 |
| abstract_inverted_index.regime-switching | 96 |
| abstract_inverted_index.simulation-based | 242 |
| abstract_inverted_index.\cite{munoz2023}; | 88 |
| abstract_inverted_index.\cite{lu2019}.Each | 126 |
| abstract_inverted_index.\cite{wattis2020}; | 109 |
| abstract_inverted_index.telomere-senescence | 198 |
| abstract_inverted_index.\cite{riihimaki2010, | 211 |
| abstract_inverted_index.GP-HMM-SDE-epigenetic | 195 |
| abstract_inverted_index.\cite{riihimaki2010}; | 70 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 1 |
| citation_normalized_percentile |