Epigenetic priming of mammalian embryonic enhancer elements coordinates developmental gene networks Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1186/s13059-025-03658-8
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1186/s13059-025-03658-8
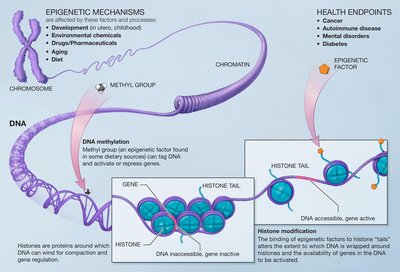
Background Embryonic development requires the accurate spatiotemporal execution of cell lineage-specific gene expression programs, which are controlled by transcriptional enhancers. Developmental enhancers adopt a primed chromatin state prior to their activation. How this primed enhancer state is established and maintained and how it affects the regulation of developmental gene networks remains poorly understood. Results Here, we use comparative multi-omic analyses of human and mouse early embryonic development to identify subsets of postgastrulation lineage-specific enhancers which are epigenetically primed ahead of their activation, marked by the histone modification H3K4me1 within the epiblast. We show that epigenetic priming occurs at lineage-specific enhancers for all three germ layers and that epigenetic priming of enhancers confers lineage-specific regulation of key developmental gene networks. Surprisingly in some cases, lineage-specific enhancers are epigenetically marked already in the zygote, weeks before their activation during lineage specification. Moreover, we outline a generalizable strategy to use naturally occurring human genetic variation to delineate important sequence determinants of primed enhancer function. Conclusions Our findings identify an evolutionarily conserved program of enhancer priming and begin to dissect the temporal dynamics and mechanisms of its establishment and maintenance during early mammalian development.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1186/s13059-025-03658-8
- OA Status
- gold
- Cited By
- 3
- References
- 82
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4412522182
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4412522182Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1186/s13059-025-03658-8Digital Object Identifier
- Title
-
Epigenetic priming of mammalian embryonic enhancer elements coordinates developmental gene networksWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-07-18Full publication date if available
- Authors
-
Christopher D Todd, Jannat Ijaz, Fereshteh Torabi, Oleksandr Dovgusha, Steve Bevan, Olivia Cracknell, Tim Lohoff, Stephen J. Clark, Ricard Argelaguet, Joshua L. Pierce, Ioannis Kafetzopoulos, Alice Santambrogio, Jennifer Nichols, Ferdinand von Meyenn, Ufuk Günesdogan, Stefan Schoenfelder, Wolf ReikList of authors in order
- Landing page
-
https://doi.org/10.1186/s13059-025-03658-8Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1186/s13059-025-03658-8Direct OA link when available
- Concepts
-
Enhancer, Biology, Epigenetics, Chromatin, Genetics, Lineage (genetic), Histone, Regulation of gene expression, Gene, Enhancer RNAs, Cell biology, Evolutionary biology, Gene expressionTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
3Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3Per-year citation counts (last 5 years)
- References (count)
-
82Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4412522182 |
|---|---|
| doi | https://doi.org/10.1186/s13059-025-03658-8 |
| ids.doi | https://doi.org/10.1186/s13059-025-03658-8 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40682180 |
| ids.openalex | https://openalex.org/W4412522182 |
| fwci | 8.04883388 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D004742 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Enhancer Elements, Genetic |
| mesh[1].qualifier_ui | |
| mesh[1].descriptor_ui | D000818 |
| mesh[1].is_major_topic | False |
| mesh[1].qualifier_name | |
| mesh[1].descriptor_name | Animals |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D044127 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Epigenesis, Genetic |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D053263 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Gene Regulatory Networks |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D006801 |
| mesh[4].is_major_topic | False |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Humans |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D051379 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Mice |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D018507 |
| mesh[6].is_major_topic | True |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Gene Expression Regulation, Developmental |
| mesh[7].qualifier_ui | Q000235 |
| mesh[7].descriptor_ui | D047108 |
| mesh[7].is_major_topic | True |
| mesh[7].qualifier_name | genetics |
| mesh[7].descriptor_name | Embryonic Development |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D019070 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Cell Lineage |
| mesh[9].qualifier_ui | Q000378 |
| mesh[9].descriptor_ui | D006657 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | metabolism |
| mesh[9].descriptor_name | Histones |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D004742 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Enhancer Elements, Genetic |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D000818 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Animals |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D044127 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Epigenesis, Genetic |
| mesh[13].qualifier_ui | |
| mesh[13].descriptor_ui | D053263 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | |
| mesh[13].descriptor_name | Gene Regulatory Networks |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D006801 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Humans |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D051379 |
| mesh[15].is_major_topic | False |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Mice |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D018507 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Gene Expression Regulation, Developmental |
| mesh[17].qualifier_ui | Q000235 |
| mesh[17].descriptor_ui | D047108 |
| mesh[17].is_major_topic | True |
| mesh[17].qualifier_name | genetics |
| mesh[17].descriptor_name | Embryonic Development |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D019070 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Cell Lineage |
| mesh[19].qualifier_ui | Q000378 |
| mesh[19].descriptor_ui | D006657 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | metabolism |
| mesh[19].descriptor_name | Histones |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D004742 |
| mesh[20].is_major_topic | True |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Enhancer Elements, Genetic |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D000818 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Animals |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D044127 |
| mesh[22].is_major_topic | True |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Epigenesis, Genetic |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D053263 |
| mesh[23].is_major_topic | True |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Gene Regulatory Networks |
| mesh[24].qualifier_ui | |
| mesh[24].descriptor_ui | D006801 |
| mesh[24].is_major_topic | False |
| mesh[24].qualifier_name | |
| mesh[24].descriptor_name | Humans |
| mesh[25].qualifier_ui | |
| mesh[25].descriptor_ui | D051379 |
| mesh[25].is_major_topic | False |
| mesh[25].qualifier_name | |
| mesh[25].descriptor_name | Mice |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D018507 |
| mesh[26].is_major_topic | True |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | Gene Expression Regulation, Developmental |
| mesh[27].qualifier_ui | Q000235 |
| mesh[27].descriptor_ui | D047108 |
| mesh[27].is_major_topic | True |
| mesh[27].qualifier_name | genetics |
| mesh[27].descriptor_name | Embryonic Development |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D019070 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Cell Lineage |
| mesh[29].qualifier_ui | Q000378 |
| mesh[29].descriptor_ui | D006657 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | metabolism |
| mesh[29].descriptor_name | Histones |
| type | article |
| title | Epigenetic priming of mammalian embryonic enhancer elements coordinates developmental gene networks |
| biblio.issue | 1 |
| biblio.volume | 26 |
| biblio.last_page | 214 |
| biblio.first_page | 214 |
| grants[0].funder | https://openalex.org/F4320308269 |
| grants[0].award_id | Sofja Kovalevskaja Award |
| grants[0].funder_display_name | Alexander von Humboldt-Stiftung |
| grants[1].funder | https://openalex.org/F4320308269 |
| grants[1].award_id | Sofja Kovalevskaja Award |
| grants[1].funder_display_name | Alexander von Humboldt-Stiftung |
| grants[2].funder | https://openalex.org/F4320311904 |
| grants[2].award_id | 210754/Z/18/Z |
| grants[2].funder_display_name | Wellcome Trust |
| grants[3].funder | https://openalex.org/F4320314731 |
| grants[3].award_id | MR/T016787/1 |
| grants[3].funder_display_name | UK Research and Innovation |
| grants[4].funder | https://openalex.org/F4320334629 |
| grants[4].award_id | BBS/E/B/000C0421 |
| grants[4].funder_display_name | Biotechnology and Biological Sciences Research Council |
| grants[5].funder | https://openalex.org/F4320334629 |
| grants[5].award_id | BBS/E/B/000C0421 |
| grants[5].funder_display_name | Biotechnology and Biological Sciences Research Council |
| grants[6].funder | https://openalex.org/F4320334629 |
| grants[6].award_id | BBS/E/B/000C0421 |
| grants[6].funder_display_name | Biotechnology and Biological Sciences Research Council |
| grants[7].funder | https://openalex.org/F4320334678 |
| grants[7].award_id | EpiCell Linage:882798 |
| grants[7].funder_display_name | European Research Council |
| grants[8].funder | https://openalex.org/F4320334678 |
| grants[8].award_id | EpiCell Linage:882798 |
| grants[8].funder_display_name | European Research Council |
| grants[9].funder | https://openalex.org/F4320334678 |
| grants[9].award_id | EpiCell Linage:882798 |
| grants[9].funder_display_name | European Research Council |
| topics[0].id | https://openalex.org/T10222 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | Genomics and Chromatin Dynamics |
| topics[1].id | https://openalex.org/T10269 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9998000264167786 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Epigenetics and DNA Methylation |
| topics[2].id | https://openalex.org/T10878 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9961000084877014 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | CRISPR and Genetic Engineering |
| funders[0].id | https://openalex.org/F4320308269 |
| funders[0].ror | https://ror.org/012kf4317 |
| funders[0].display_name | Alexander von Humboldt-Stiftung |
| funders[1].id | https://openalex.org/F4320311904 |
| funders[1].ror | https://ror.org/029chgv08 |
| funders[1].display_name | Wellcome Trust |
| funders[2].id | https://openalex.org/F4320314731 |
| funders[2].ror | https://ror.org/001aqnf71 |
| funders[2].display_name | UK Research and Innovation |
| funders[3].id | https://openalex.org/F4320334629 |
| funders[3].ror | https://ror.org/00cwqg982 |
| funders[3].display_name | Biotechnology and Biological Sciences Research Council |
| funders[4].id | https://openalex.org/F4320334678 |
| funders[4].ror | https://ror.org/0472cxd90 |
| funders[4].display_name | European Research Council |
| is_xpac | False |
| apc_list.value | 4090 |
| apc_list.currency | EUR |
| apc_list.value_usd | 5030 |
| apc_paid.value | 4090 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 5030 |
| concepts[0].id | https://openalex.org/C111936080 |
| concepts[0].level | 4 |
| concepts[0].score | 0.9200507998466492 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q913367 |
| concepts[0].display_name | Enhancer |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.9062951803207397 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C41091548 |
| concepts[2].level | 3 |
| concepts[2].score | 0.7120425701141357 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q26939 |
| concepts[2].display_name | Epigenetics |
| concepts[3].id | https://openalex.org/C83640560 |
| concepts[3].level | 3 |
| concepts[3].score | 0.6252217888832092 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q180951 |
| concepts[3].display_name | Chromatin |
| concepts[4].id | https://openalex.org/C54355233 |
| concepts[4].level | 1 |
| concepts[4].score | 0.6038860082626343 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[4].display_name | Genetics |
| concepts[5].id | https://openalex.org/C2776817793 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5346727967262268 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q6553369 |
| concepts[5].display_name | Lineage (genetic) |
| concepts[6].id | https://openalex.org/C64927066 |
| concepts[6].level | 3 |
| concepts[6].score | 0.5041142702102661 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q36293 |
| concepts[6].display_name | Histone |
| concepts[7].id | https://openalex.org/C165864922 |
| concepts[7].level | 3 |
| concepts[7].score | 0.49951982498168945 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q411391 |
| concepts[7].display_name | Regulation of gene expression |
| concepts[8].id | https://openalex.org/C104317684 |
| concepts[8].level | 2 |
| concepts[8].score | 0.42394891381263733 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[8].display_name | Gene |
| concepts[9].id | https://openalex.org/C21784809 |
| concepts[9].level | 5 |
| concepts[9].score | 0.4163326323032379 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q17119097 |
| concepts[9].display_name | Enhancer RNAs |
| concepts[10].id | https://openalex.org/C95444343 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3558870553970337 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7141 |
| concepts[10].display_name | Cell biology |
| concepts[11].id | https://openalex.org/C78458016 |
| concepts[11].level | 1 |
| concepts[11].score | 0.33618974685668945 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q840400 |
| concepts[11].display_name | Evolutionary biology |
| concepts[12].id | https://openalex.org/C150194340 |
| concepts[12].level | 3 |
| concepts[12].score | 0.2870157063007355 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q26972 |
| concepts[12].display_name | Gene expression |
| keywords[0].id | https://openalex.org/keywords/enhancer |
| keywords[0].score | 0.9200507998466492 |
| keywords[0].display_name | Enhancer |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.9062951803207397 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/epigenetics |
| keywords[2].score | 0.7120425701141357 |
| keywords[2].display_name | Epigenetics |
| keywords[3].id | https://openalex.org/keywords/chromatin |
| keywords[3].score | 0.6252217888832092 |
| keywords[3].display_name | Chromatin |
| keywords[4].id | https://openalex.org/keywords/genetics |
| keywords[4].score | 0.6038860082626343 |
| keywords[4].display_name | Genetics |
| keywords[5].id | https://openalex.org/keywords/lineage |
| keywords[5].score | 0.5346727967262268 |
| keywords[5].display_name | Lineage (genetic) |
| keywords[6].id | https://openalex.org/keywords/histone |
| keywords[6].score | 0.5041142702102661 |
| keywords[6].display_name | Histone |
| keywords[7].id | https://openalex.org/keywords/regulation-of-gene-expression |
| keywords[7].score | 0.49951982498168945 |
| keywords[7].display_name | Regulation of gene expression |
| keywords[8].id | https://openalex.org/keywords/gene |
| keywords[8].score | 0.42394891381263733 |
| keywords[8].display_name | Gene |
| keywords[9].id | https://openalex.org/keywords/enhancer-rnas |
| keywords[9].score | 0.4163326323032379 |
| keywords[9].display_name | Enhancer RNAs |
| keywords[10].id | https://openalex.org/keywords/cell-biology |
| keywords[10].score | 0.3558870553970337 |
| keywords[10].display_name | Cell biology |
| keywords[11].id | https://openalex.org/keywords/evolutionary-biology |
| keywords[11].score | 0.33618974685668945 |
| keywords[11].display_name | Evolutionary biology |
| keywords[12].id | https://openalex.org/keywords/gene-expression |
| keywords[12].score | 0.2870157063007355 |
| keywords[12].display_name | Gene expression |
| language | en |
| locations[0].id | doi:10.1186/s13059-025-03658-8 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S81160022 |
| locations[0].source.issn | 1474-7596, 1474-760X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1474-7596 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Genome biology |
| locations[0].source.host_organization | https://openalex.org/P4310320256 |
| locations[0].source.host_organization_name | BioMed Central |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320256 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Genome Biology |
| locations[0].landing_page_url | https://doi.org/10.1186/s13059-025-03658-8 |
| locations[1].id | pmid:40682180 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Genome biology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40682180 |
| locations[2].id | pmh:oai:publications.goettingen-research-online.de:2/149855 |
| locations[2].is_oa | False |
| locations[2].source | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | journal_article |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://resolver.sub.uni-goettingen.de/purl?gro-2/149855 |
| locations[3].id | pmh:oai:doaj.org/article:8d9d70d62143447daf2bd99f30d54596 |
| locations[3].is_oa | False |
| locations[3].source.id | https://openalex.org/S4306401280 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[3].source.host_organization | |
| locations[3].source.host_organization_name | |
| locations[3].source.host_organization_lineage | |
| locations[3].license | |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | article |
| locations[3].license_id | |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | Genome Biology, Vol 26, Iss 1, Pp 1-23 (2025) |
| locations[3].landing_page_url | https://doaj.org/article/8d9d70d62143447daf2bd99f30d54596 |
| locations[4].id | pmh:oai:pubmedcentral.nih.gov:12272991 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S2764455111 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | PubMed Central |
| locations[4].source.host_organization | https://openalex.org/I1299303238 |
| locations[4].source.host_organization_name | National Institutes of Health |
| locations[4].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[4].license | other-oa |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/other-oa |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Genome Biol |
| locations[4].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/12272991 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5081873417 |
| authorships[0].author.orcid | https://orcid.org/0000-0003-2663-6173 |
| authorships[0].author.display_name | Christopher D Todd |
| authorships[0].countries | GB |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[0].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. [email protected]. |
| authorships[0].affiliations[1].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. [email protected]. |
| authorships[0].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[0].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[0].institutions[0].country_code | GB |
| authorships[0].institutions[0].display_name | Babraham Institute |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Christopher D Todd |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. [email protected]., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. [email protected]. |
| authorships[1].author.id | https://openalex.org/A5065346983 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-9034-9983 |
| authorships[1].author.display_name | Jannat Ijaz |
| authorships[1].countries | GB |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210110691 |
| authorships[1].affiliations[0].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. |
| authorships[1].affiliations[1].institution_ids | https://openalex.org/I1302113299 |
| authorships[1].affiliations[1].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[1].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[1].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[1].institutions[0].country_code | GB |
| authorships[1].institutions[0].display_name | Babraham Institute |
| authorships[1].institutions[1].id | https://openalex.org/I4210110691 |
| authorships[1].institutions[1].ror | https://ror.org/01x5y8335 |
| authorships[1].institutions[1].type | company |
| authorships[1].institutions[1].lineage | https://openalex.org/I4210110691 |
| authorships[1].institutions[1].country_code | GB |
| authorships[1].institutions[1].display_name | Granta Design (United Kingdom) |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Jannat Ijaz |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[2].author.id | https://openalex.org/A5013104202 |
| authorships[2].author.orcid | https://orcid.org/0009-0008-8660-3350 |
| authorships[2].author.display_name | Fereshteh Torabi |
| authorships[2].countries | GB |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I2802476451 |
| authorships[2].affiliations[0].raw_affiliation_string | Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, UK. |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I1302113299 |
| authorships[2].affiliations[1].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[2].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[2].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[2].institutions[0].type | facility |
| authorships[2].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[2].institutions[0].country_code | GB |
| authorships[2].institutions[0].display_name | Babraham Institute |
| authorships[2].institutions[1].id | https://openalex.org/I2802476451 |
| authorships[2].institutions[1].ror | https://ror.org/05cy4wa09 |
| authorships[2].institutions[1].type | nonprofit |
| authorships[2].institutions[1].lineage | https://openalex.org/I2802476451, https://openalex.org/I87048295 |
| authorships[2].institutions[1].country_code | GB |
| authorships[2].institutions[1].display_name | Wellcome Sanger Institute |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Fereshteh Torabi |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK., Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, UK. |
| authorships[3].author.id | https://openalex.org/A5043486224 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-4846-9409 |
| authorships[3].author.display_name | Oleksandr Dovgusha |
| authorships[3].countries | DE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I74656192 |
| authorships[3].affiliations[0].raw_affiliation_string | Göttingen Center for Molecular Biology, Department of Developmental Biology, University of Göttingen, Justus-Von-Liebig Weg 11, Göttingen, 37077, Germany. |
| authorships[3].institutions[0].id | https://openalex.org/I74656192 |
| authorships[3].institutions[0].ror | https://ror.org/01y9bpm73 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I74656192 |
| authorships[3].institutions[0].country_code | DE |
| authorships[3].institutions[0].display_name | University of Göttingen |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Oleksandr Dovgusha |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Göttingen Center for Molecular Biology, Department of Developmental Biology, University of Göttingen, Justus-Von-Liebig Weg 11, Göttingen, 37077, Germany. |
| authorships[4].author.id | https://openalex.org/A5049405504 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-0490-6830 |
| authorships[4].author.display_name | Steve Bevan |
| authorships[4].countries | GB |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[4].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I2800110054 |
| authorships[4].affiliations[1].raw_affiliation_string | GSK, Gunnels Wood Road, Stevenage, SG1 2NY, UK. |
| authorships[4].institutions[0].id | https://openalex.org/I2800110054 |
| authorships[4].institutions[0].ror | https://ror.org/050x9d346 |
| authorships[4].institutions[0].type | nonprofit |
| authorships[4].institutions[0].lineage | https://openalex.org/I2800110054 |
| authorships[4].institutions[0].country_code | GB |
| authorships[4].institutions[0].display_name | Age UK |
| authorships[4].institutions[1].id | https://openalex.org/I1302113299 |
| authorships[4].institutions[1].ror | https://ror.org/01d5qpn59 |
| authorships[4].institutions[1].type | facility |
| authorships[4].institutions[1].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[4].institutions[1].country_code | GB |
| authorships[4].institutions[1].display_name | Babraham Institute |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Stephen Bevan |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK., GSK, Gunnels Wood Road, Stevenage, SG1 2NY, UK. |
| authorships[5].author.id | https://openalex.org/A5107109258 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Olivia Cracknell |
| authorships[5].countries | GB |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[5].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[5].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[5].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[5].institutions[0].type | facility |
| authorships[5].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[5].institutions[0].country_code | GB |
| authorships[5].institutions[0].display_name | Babraham Institute |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Olivia Cracknell |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[6].author.id | https://openalex.org/A5001445617 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-9333-842X |
| authorships[6].author.display_name | Tim Lohoff |
| authorships[6].countries | GB |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[6].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[6].affiliations[1].raw_affiliation_string | Forbion, Gaertnerplatz 6, Munich, Germany. |
| authorships[6].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[6].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[6].institutions[0].type | facility |
| authorships[6].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[6].institutions[0].country_code | GB |
| authorships[6].institutions[0].display_name | Babraham Institute |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Tim Lohoff |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK., Forbion, Gaertnerplatz 6, Munich, Germany. |
| authorships[7].author.id | https://openalex.org/A5091359674 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-6183-491X |
| authorships[7].author.display_name | Stephen J. Clark |
| authorships[7].countries | GB |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[7].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I4210110691 |
| authorships[7].affiliations[1].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. |
| authorships[7].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[7].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[7].institutions[0].country_code | GB |
| authorships[7].institutions[0].display_name | Babraham Institute |
| authorships[7].institutions[1].id | https://openalex.org/I4210110691 |
| authorships[7].institutions[1].ror | https://ror.org/01x5y8335 |
| authorships[7].institutions[1].type | company |
| authorships[7].institutions[1].lineage | https://openalex.org/I4210110691 |
| authorships[7].institutions[1].country_code | GB |
| authorships[7].institutions[1].display_name | Granta Design (United Kingdom) |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Stephen Clark |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[8].author.id | https://openalex.org/A5005275806 |
| authorships[8].author.orcid | https://orcid.org/0000-0003-3199-3722 |
| authorships[8].author.display_name | Ricard Argelaguet |
| authorships[8].countries | GB |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[8].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[8].affiliations[1].institution_ids | https://openalex.org/I4210110691 |
| authorships[8].affiliations[1].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. |
| authorships[8].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[8].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[8].institutions[0].type | facility |
| authorships[8].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[8].institutions[0].country_code | GB |
| authorships[8].institutions[0].display_name | Babraham Institute |
| authorships[8].institutions[1].id | https://openalex.org/I4210110691 |
| authorships[8].institutions[1].ror | https://ror.org/01x5y8335 |
| authorships[8].institutions[1].type | company |
| authorships[8].institutions[1].lineage | https://openalex.org/I4210110691 |
| authorships[8].institutions[1].country_code | GB |
| authorships[8].institutions[1].display_name | Granta Design (United Kingdom) |
| authorships[8].author_position | middle |
| authorships[8].raw_author_name | Ricard Argelaguet |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[9].author.id | https://openalex.org/A5002154225 |
| authorships[9].author.orcid | https://orcid.org/0000-0002-2773-6793 |
| authorships[9].author.display_name | Joshua L. Pierce |
| authorships[9].countries | DE, GB |
| authorships[9].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[9].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[9].affiliations[1].institution_ids | https://openalex.org/I4210120689, https://openalex.org/I887968799 |
| authorships[9].affiliations[1].raw_affiliation_string | Institute for Cell and Tumor Biology, RWTH Aachen University Hospital, Aachen, Germany. |
| authorships[9].institutions[0].id | https://openalex.org/I887968799 |
| authorships[9].institutions[0].ror | https://ror.org/04xfq0f34 |
| authorships[9].institutions[0].type | education |
| authorships[9].institutions[0].lineage | https://openalex.org/I887968799 |
| authorships[9].institutions[0].country_code | DE |
| authorships[9].institutions[0].display_name | RWTH Aachen University |
| authorships[9].institutions[1].id | https://openalex.org/I4210120689 |
| authorships[9].institutions[1].ror | https://ror.org/02gm5zw39 |
| authorships[9].institutions[1].type | healthcare |
| authorships[9].institutions[1].lineage | https://openalex.org/I4210120689 |
| authorships[9].institutions[1].country_code | DE |
| authorships[9].institutions[1].display_name | Universitätsklinikum Aachen |
| authorships[9].institutions[2].id | https://openalex.org/I1302113299 |
| authorships[9].institutions[2].ror | https://ror.org/01d5qpn59 |
| authorships[9].institutions[2].type | facility |
| authorships[9].institutions[2].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[9].institutions[2].country_code | GB |
| authorships[9].institutions[2].display_name | Babraham Institute |
| authorships[9].author_position | middle |
| authorships[9].raw_author_name | Juliette Pierce |
| authorships[9].is_corresponding | False |
| authorships[9].raw_affiliation_strings | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK., Institute for Cell and Tumor Biology, RWTH Aachen University Hospital, Aachen, Germany. |
| authorships[10].author.id | https://openalex.org/A5062358164 |
| authorships[10].author.orcid | https://orcid.org/0000-0001-5948-3455 |
| authorships[10].author.display_name | Ioannis Kafetzopoulos |
| authorships[10].countries | GB |
| authorships[10].affiliations[0].institution_ids | https://openalex.org/I4210110691 |
| authorships[10].affiliations[0].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. |
| authorships[10].affiliations[1].institution_ids | https://openalex.org/I1302113299 |
| authorships[10].affiliations[1].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[10].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[10].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[10].institutions[0].type | facility |
| authorships[10].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[10].institutions[0].country_code | GB |
| authorships[10].institutions[0].display_name | Babraham Institute |
| authorships[10].institutions[1].id | https://openalex.org/I4210110691 |
| authorships[10].institutions[1].ror | https://ror.org/01x5y8335 |
| authorships[10].institutions[1].type | company |
| authorships[10].institutions[1].lineage | https://openalex.org/I4210110691 |
| authorships[10].institutions[1].country_code | GB |
| authorships[10].institutions[1].display_name | Granta Design (United Kingdom) |
| authorships[10].author_position | middle |
| authorships[10].raw_author_name | Ioannis Kafetzopoulos |
| authorships[10].is_corresponding | False |
| authorships[10].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[11].author.id | https://openalex.org/A5058737503 |
| authorships[11].author.orcid | https://orcid.org/0000-0001-6829-8622 |
| authorships[11].author.display_name | Alice Santambrogio |
| authorships[11].countries | GB |
| authorships[11].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[11].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[11].affiliations[1].institution_ids | https://openalex.org/I4210110691 |
| authorships[11].affiliations[1].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. |
| authorships[11].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[11].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[11].institutions[0].type | facility |
| authorships[11].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[11].institutions[0].country_code | GB |
| authorships[11].institutions[0].display_name | Babraham Institute |
| authorships[11].institutions[1].id | https://openalex.org/I4210110691 |
| authorships[11].institutions[1].ror | https://ror.org/01x5y8335 |
| authorships[11].institutions[1].type | company |
| authorships[11].institutions[1].lineage | https://openalex.org/I4210110691 |
| authorships[11].institutions[1].country_code | GB |
| authorships[11].institutions[1].display_name | Granta Design (United Kingdom) |
| authorships[11].author_position | middle |
| authorships[11].raw_author_name | Alice Santambrogio |
| authorships[11].is_corresponding | False |
| authorships[11].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[12].author.id | https://openalex.org/A5008565592 |
| authorships[12].author.orcid | https://orcid.org/0000-0002-8650-1388 |
| authorships[12].author.display_name | Jennifer Nichols |
| authorships[12].countries | GB |
| authorships[12].affiliations[0].institution_ids | https://openalex.org/I21196054, https://openalex.org/I241749 |
| authorships[12].affiliations[0].raw_affiliation_string | Wellcome-MRC Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre, University of Cambridge, Puddicombe Way, Cambridge, CB2 0AW, UK. |
| authorships[12].affiliations[1].institution_ids | https://openalex.org/I2800150677, https://openalex.org/I98677209 |
| authorships[12].affiliations[1].raw_affiliation_string | Institute of Genetics and Cancer, The University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, UK. |
| authorships[12].institutions[0].id | https://openalex.org/I241749 |
| authorships[12].institutions[0].ror | https://ror.org/013meh722 |
| authorships[12].institutions[0].type | education |
| authorships[12].institutions[0].lineage | https://openalex.org/I241749 |
| authorships[12].institutions[0].country_code | GB |
| authorships[12].institutions[0].display_name | University of Cambridge |
| authorships[12].institutions[1].id | https://openalex.org/I98677209 |
| authorships[12].institutions[1].ror | https://ror.org/01nrxwf90 |
| authorships[12].institutions[1].type | education |
| authorships[12].institutions[1].lineage | https://openalex.org/I98677209 |
| authorships[12].institutions[1].country_code | GB |
| authorships[12].institutions[1].display_name | University of Edinburgh |
| authorships[12].institutions[2].id | https://openalex.org/I21196054 |
| authorships[12].institutions[2].ror | https://ror.org/05nz0zp31 |
| authorships[12].institutions[2].type | facility |
| authorships[12].institutions[2].lineage | https://openalex.org/I21196054, https://openalex.org/I241749, https://openalex.org/I4210087105, https://openalex.org/I87048295, https://openalex.org/I90344618 |
| authorships[12].institutions[2].country_code | GB |
| authorships[12].institutions[2].display_name | Wellcome/MRC Cambridge Stem Cell Institute |
| authorships[12].institutions[3].id | https://openalex.org/I2800150677 |
| authorships[12].institutions[3].ror | https://ror.org/009kr6r15 |
| authorships[12].institutions[3].type | healthcare |
| authorships[12].institutions[3].lineage | https://openalex.org/I1324252669, https://openalex.org/I2800150677 |
| authorships[12].institutions[3].country_code | GB |
| authorships[12].institutions[3].display_name | Western General Hospital |
| authorships[12].author_position | middle |
| authorships[12].raw_author_name | Jennifer Nichols |
| authorships[12].is_corresponding | False |
| authorships[12].raw_affiliation_strings | Institute of Genetics and Cancer, The University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, UK., Wellcome-MRC Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre, University of Cambridge, Puddicombe Way, Cambridge, CB2 0AW, UK. |
| authorships[13].author.id | https://openalex.org/A5005404209 |
| authorships[13].author.orcid | https://orcid.org/0000-0001-9920-3075 |
| authorships[13].author.display_name | Ferdinand von Meyenn |
| authorships[13].countries | CH, GB |
| authorships[13].affiliations[0].institution_ids | https://openalex.org/I183935753 |
| authorships[13].affiliations[0].raw_affiliation_string | Department of Medical and Molecular Genetics, Kings College London, London, UK. |
| authorships[13].affiliations[1].institution_ids | https://openalex.org/I1302113299 |
| authorships[13].affiliations[1].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. |
| authorships[13].affiliations[2].institution_ids | https://openalex.org/I35440088 |
| authorships[13].affiliations[2].raw_affiliation_string | Laboratory of Nutrition and Metabolic Epigenetics, Department of Health Sciences and Technology, ETH Zurich, Zurich, Switzerland. |
| authorships[13].institutions[0].id | https://openalex.org/I35440088 |
| authorships[13].institutions[0].ror | https://ror.org/05a28rw58 |
| authorships[13].institutions[0].type | education |
| authorships[13].institutions[0].lineage | https://openalex.org/I2799323385, https://openalex.org/I35440088 |
| authorships[13].institutions[0].country_code | CH |
| authorships[13].institutions[0].display_name | ETH Zurich |
| authorships[13].institutions[1].id | https://openalex.org/I1302113299 |
| authorships[13].institutions[1].ror | https://ror.org/01d5qpn59 |
| authorships[13].institutions[1].type | facility |
| authorships[13].institutions[1].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[13].institutions[1].country_code | GB |
| authorships[13].institutions[1].display_name | Babraham Institute |
| authorships[13].institutions[2].id | https://openalex.org/I183935753 |
| authorships[13].institutions[2].ror | https://ror.org/0220mzb33 |
| authorships[13].institutions[2].type | education |
| authorships[13].institutions[2].lineage | https://openalex.org/I124357947, https://openalex.org/I183935753 |
| authorships[13].institutions[2].country_code | GB |
| authorships[13].institutions[2].display_name | King's College London |
| authorships[13].author_position | middle |
| authorships[13].raw_author_name | Ferdinand von Meyenn |
| authorships[13].is_corresponding | False |
| authorships[13].raw_affiliation_strings | Department of Medical and Molecular Genetics, Kings College London, London, UK., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK., Laboratory of Nutrition and Metabolic Epigenetics, Department of Health Sciences and Technology, ETH Zurich, Zurich, Switzerland. |
| authorships[14].author.id | https://openalex.org/A5079142953 |
| authorships[14].author.orcid | https://orcid.org/0000-0002-8658-8022 |
| authorships[14].author.display_name | Ufuk Günesdogan |
| authorships[14].countries | DE |
| authorships[14].affiliations[0].institution_ids | https://openalex.org/I74656192 |
| authorships[14].affiliations[0].raw_affiliation_string | Göttingen Center for Molecular Biology, Department of Developmental Biology, University of Göttingen, Justus-Von-Liebig Weg 11, Göttingen, 37077, Germany. |
| authorships[14].institutions[0].id | https://openalex.org/I74656192 |
| authorships[14].institutions[0].ror | https://ror.org/01y9bpm73 |
| authorships[14].institutions[0].type | education |
| authorships[14].institutions[0].lineage | https://openalex.org/I74656192 |
| authorships[14].institutions[0].country_code | DE |
| authorships[14].institutions[0].display_name | University of Göttingen |
| authorships[14].author_position | middle |
| authorships[14].raw_author_name | Ufuk Günesdogan |
| authorships[14].is_corresponding | False |
| authorships[14].raw_affiliation_strings | Göttingen Center for Molecular Biology, Department of Developmental Biology, University of Göttingen, Justus-Von-Liebig Weg 11, Göttingen, 37077, Germany. |
| authorships[15].author.id | https://openalex.org/A5090153306 |
| authorships[15].author.orcid | https://orcid.org/0000-0002-3200-8133 |
| authorships[15].author.display_name | Stefan Schoenfelder |
| authorships[15].countries | GB |
| authorships[15].affiliations[0].institution_ids | https://openalex.org/I1302113299 |
| authorships[15].affiliations[0].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. [email protected]. |
| authorships[15].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[15].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[15].institutions[0].type | facility |
| authorships[15].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[15].institutions[0].country_code | GB |
| authorships[15].institutions[0].display_name | Babraham Institute |
| authorships[15].author_position | middle |
| authorships[15].raw_author_name | Stefan Schoenfelder |
| authorships[15].is_corresponding | False |
| authorships[15].raw_affiliation_strings | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. [email protected]. |
| authorships[16].author.id | https://openalex.org/A5052074743 |
| authorships[16].author.orcid | https://orcid.org/0000-0003-0216-9881 |
| authorships[16].author.display_name | Wolf Reik |
| authorships[16].countries | GB |
| authorships[16].affiliations[0].raw_affiliation_string | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. [email protected]. |
| authorships[16].affiliations[1].institution_ids | https://openalex.org/I1302113299 |
| authorships[16].affiliations[1].raw_affiliation_string | Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. [email protected]. |
| authorships[16].institutions[0].id | https://openalex.org/I1302113299 |
| authorships[16].institutions[0].ror | https://ror.org/01d5qpn59 |
| authorships[16].institutions[0].type | facility |
| authorships[16].institutions[0].lineage | https://openalex.org/I1302113299, https://openalex.org/I2799693246, https://openalex.org/I4210087105 |
| authorships[16].institutions[0].country_code | GB |
| authorships[16].institutions[0].display_name | Babraham Institute |
| authorships[16].author_position | last |
| authorships[16].raw_author_name | Wolf Reik |
| authorships[16].is_corresponding | False |
| authorships[16].raw_affiliation_strings | Altos Labs, Cambridge Institute of Science, Granta Park, Cambridge, CB21 6GQ, UK. [email protected]., Epigenetics Programme, Babraham Institute, Babraham Research Campus, Cambridge, CB22 3AT, UK. [email protected]. |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1186/s13059-025-03658-8 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Epigenetic priming of mammalian embryonic enhancer elements coordinates developmental gene networks |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10222 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | Genomics and Chromatin Dynamics |
| related_works | https://openalex.org/W2113330652, https://openalex.org/W3098554796, https://openalex.org/W2567553314, https://openalex.org/W3069949010, https://openalex.org/W2146648243, https://openalex.org/W2010420636, https://openalex.org/W4236711043, https://openalex.org/W2623141677, https://openalex.org/W2992086063, https://openalex.org/W2688223077 |
| cited_by_count | 3 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| locations_count | 5 |
| best_oa_location.id | doi:10.1186/s13059-025-03658-8 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S81160022 |
| best_oa_location.source.issn | 1474-7596, 1474-760X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1474-7596 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Genome biology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320256 |
| best_oa_location.source.host_organization_name | BioMed Central |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320256 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Genome Biology |
| best_oa_location.landing_page_url | https://doi.org/10.1186/s13059-025-03658-8 |
| primary_location.id | doi:10.1186/s13059-025-03658-8 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S81160022 |
| primary_location.source.issn | 1474-7596, 1474-760X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1474-7596 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Genome biology |
| primary_location.source.host_organization | https://openalex.org/P4310320256 |
| primary_location.source.host_organization_name | BioMed Central |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320256 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Genome Biology |
| primary_location.landing_page_url | https://doi.org/10.1186/s13059-025-03658-8 |
| publication_date | 2025-07-18 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W3131055609, https://openalex.org/W2519629804, https://openalex.org/W2131374955, https://openalex.org/W2800074119, https://openalex.org/W3033596656, https://openalex.org/W2076154138, https://openalex.org/W4315621830, https://openalex.org/W2988510233, https://openalex.org/W3103797259, https://openalex.org/W3108971791, https://openalex.org/W2171808845, https://openalex.org/W2119180969, https://openalex.org/W2341539131, https://openalex.org/W2129785061, https://openalex.org/W2036897871, https://openalex.org/W4402417773, https://openalex.org/W1981590299, https://openalex.org/W4308834893, https://openalex.org/W2920191405, https://openalex.org/W2613179652, https://openalex.org/W2014677321, https://openalex.org/W4399735060, https://openalex.org/W2145144631, https://openalex.org/W1991655646, https://openalex.org/W2465074434, https://openalex.org/W1986586739, https://openalex.org/W2988126442, https://openalex.org/W2098211567, https://openalex.org/W2259938310, https://openalex.org/W2793965282, https://openalex.org/W2626695356, https://openalex.org/W2602710346, https://openalex.org/W2154746182, https://openalex.org/W3007859856, https://openalex.org/W2530086808, https://openalex.org/W2117757143, https://openalex.org/W4313826374, https://openalex.org/W2170551349, https://openalex.org/W4323042817, https://openalex.org/W2048350099, https://openalex.org/W2163732711, https://openalex.org/W2102619694, https://openalex.org/W1997453048, https://openalex.org/W2996429202, https://openalex.org/W2159164310, https://openalex.org/W2951674772, https://openalex.org/W2167776710, https://openalex.org/W2108664872, https://openalex.org/W2594685900, https://openalex.org/W2163811581, https://openalex.org/W3047447368, https://openalex.org/W2053433605, https://openalex.org/W2470453245, https://openalex.org/W2019005509, https://openalex.org/W2528171814, https://openalex.org/W2299358699, https://openalex.org/W1988568961, https://openalex.org/W2992957227, https://openalex.org/W4294309410, https://openalex.org/W2923571344, https://openalex.org/W2750416916, https://openalex.org/W2955718886, https://openalex.org/W2157232603, https://openalex.org/W2777639606, https://openalex.org/W2796674913, https://openalex.org/W4225567851, https://openalex.org/W2956942305, https://openalex.org/W4244258249, https://openalex.org/W2058613034, https://openalex.org/W2061372269, https://openalex.org/W2060446375, https://openalex.org/W1755674448, https://openalex.org/W1981463058, https://openalex.org/W2743316243, https://openalex.org/W2075928340, https://openalex.org/W3150031743, https://openalex.org/W1988036288, https://openalex.org/W2008227324, https://openalex.org/W2020696713, https://openalex.org/W1981030303, https://openalex.org/W2123564569, https://openalex.org/W2033201031 |
| referenced_works_count | 82 |
| abstract_inverted_index.a | 24, 143 |
| abstract_inverted_index.We | 92 |
| abstract_inverted_index.an | 166 |
| abstract_inverted_index.at | 98 |
| abstract_inverted_index.by | 18, 84 |
| abstract_inverted_index.in | 121, 130 |
| abstract_inverted_index.is | 37 |
| abstract_inverted_index.it | 43 |
| abstract_inverted_index.of | 9, 47, 61, 71, 80, 110, 115, 158, 170, 182 |
| abstract_inverted_index.to | 29, 68, 146, 153, 175 |
| abstract_inverted_index.we | 56, 141 |
| abstract_inverted_index.How | 32 |
| abstract_inverted_index.Our | 163 |
| abstract_inverted_index.all | 102 |
| abstract_inverted_index.and | 39, 41, 63, 106, 173, 180, 185 |
| abstract_inverted_index.are | 16, 76, 126 |
| abstract_inverted_index.for | 101 |
| abstract_inverted_index.how | 42 |
| abstract_inverted_index.its | 183 |
| abstract_inverted_index.key | 116 |
| abstract_inverted_index.the | 5, 45, 85, 90, 131, 177 |
| abstract_inverted_index.use | 57, 147 |
| abstract_inverted_index.cell | 10 |
| abstract_inverted_index.gene | 12, 49, 118 |
| abstract_inverted_index.germ | 104 |
| abstract_inverted_index.show | 93 |
| abstract_inverted_index.some | 122 |
| abstract_inverted_index.that | 94, 107 |
| abstract_inverted_index.this | 33 |
| abstract_inverted_index.Here, | 55 |
| abstract_inverted_index.adopt | 23 |
| abstract_inverted_index.ahead | 79 |
| abstract_inverted_index.begin | 174 |
| abstract_inverted_index.early | 65, 188 |
| abstract_inverted_index.human | 62, 150 |
| abstract_inverted_index.mouse | 64 |
| abstract_inverted_index.prior | 28 |
| abstract_inverted_index.state | 27, 36 |
| abstract_inverted_index.their | 30, 81, 135 |
| abstract_inverted_index.three | 103 |
| abstract_inverted_index.weeks | 133 |
| abstract_inverted_index.which | 15, 75 |
| abstract_inverted_index.before | 134 |
| abstract_inverted_index.cases, | 123 |
| abstract_inverted_index.during | 137, 187 |
| abstract_inverted_index.layers | 105 |
| abstract_inverted_index.marked | 83, 128 |
| abstract_inverted_index.occurs | 97 |
| abstract_inverted_index.poorly | 52 |
| abstract_inverted_index.primed | 25, 34, 78, 159 |
| abstract_inverted_index.within | 89 |
| abstract_inverted_index.H3K4me1 | 88 |
| abstract_inverted_index.Results | 54 |
| abstract_inverted_index.affects | 44 |
| abstract_inverted_index.already | 129 |
| abstract_inverted_index.confers | 112 |
| abstract_inverted_index.dissect | 176 |
| abstract_inverted_index.genetic | 151 |
| abstract_inverted_index.histone | 86 |
| abstract_inverted_index.lineage | 138 |
| abstract_inverted_index.outline | 142 |
| abstract_inverted_index.priming | 96, 109, 172 |
| abstract_inverted_index.program | 169 |
| abstract_inverted_index.remains | 51 |
| abstract_inverted_index.subsets | 70 |
| abstract_inverted_index.zygote, | 132 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.accurate | 6 |
| abstract_inverted_index.analyses | 60 |
| abstract_inverted_index.dynamics | 179 |
| abstract_inverted_index.enhancer | 35, 160, 171 |
| abstract_inverted_index.findings | 164 |
| abstract_inverted_index.identify | 69, 165 |
| abstract_inverted_index.networks | 50 |
| abstract_inverted_index.requires | 4 |
| abstract_inverted_index.sequence | 156 |
| abstract_inverted_index.strategy | 145 |
| abstract_inverted_index.temporal | 178 |
| abstract_inverted_index.Embryonic | 2 |
| abstract_inverted_index.Moreover, | 140 |
| abstract_inverted_index.chromatin | 26 |
| abstract_inverted_index.conserved | 168 |
| abstract_inverted_index.delineate | 154 |
| abstract_inverted_index.embryonic | 66 |
| abstract_inverted_index.enhancers | 22, 74, 100, 111, 125 |
| abstract_inverted_index.epiblast. | 91 |
| abstract_inverted_index.execution | 8 |
| abstract_inverted_index.function. | 161 |
| abstract_inverted_index.important | 155 |
| abstract_inverted_index.mammalian | 189 |
| abstract_inverted_index.naturally | 148 |
| abstract_inverted_index.networks. | 119 |
| abstract_inverted_index.occurring | 149 |
| abstract_inverted_index.programs, | 14 |
| abstract_inverted_index.variation | 152 |
| abstract_inverted_index.Background | 1 |
| abstract_inverted_index.activation | 136 |
| abstract_inverted_index.controlled | 17 |
| abstract_inverted_index.enhancers. | 20 |
| abstract_inverted_index.epigenetic | 95, 108 |
| abstract_inverted_index.expression | 13 |
| abstract_inverted_index.maintained | 40 |
| abstract_inverted_index.mechanisms | 181 |
| abstract_inverted_index.multi-omic | 59 |
| abstract_inverted_index.regulation | 46, 114 |
| abstract_inverted_index.Conclusions | 162 |
| abstract_inverted_index.activation, | 82 |
| abstract_inverted_index.activation. | 31 |
| abstract_inverted_index.comparative | 58 |
| abstract_inverted_index.development | 3, 67 |
| abstract_inverted_index.established | 38 |
| abstract_inverted_index.maintenance | 186 |
| abstract_inverted_index.understood. | 53 |
| abstract_inverted_index.Surprisingly | 120 |
| abstract_inverted_index.determinants | 157 |
| abstract_inverted_index.development. | 190 |
| abstract_inverted_index.modification | 87 |
| abstract_inverted_index.Developmental | 21 |
| abstract_inverted_index.developmental | 48, 117 |
| abstract_inverted_index.establishment | 184 |
| abstract_inverted_index.generalizable | 144 |
| abstract_inverted_index.epigenetically | 77, 127 |
| abstract_inverted_index.evolutionarily | 167 |
| abstract_inverted_index.spatiotemporal | 7 |
| abstract_inverted_index.specification. | 139 |
| abstract_inverted_index.transcriptional | 19 |
| abstract_inverted_index.lineage-specific | 11, 73, 99, 113, 124 |
| abstract_inverted_index.postgastrulation | 72 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 96 |
| countries_distinct_count | 3 |
| institutions_distinct_count | 17 |
| citation_normalized_percentile.value | 0.94396357 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |