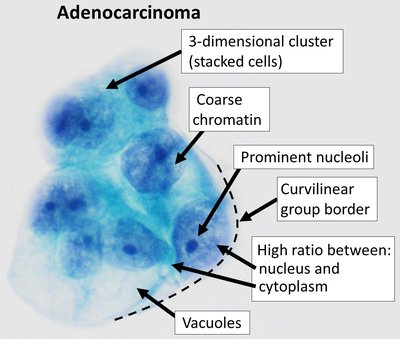
Evaluating generative AI models for explainable pathological feature extraction in lung adenocarcinoma: grading assessment and prognostic model construction Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1016/j.lanwpc.2024.101352
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1016/j.lanwpc.2024.101352
Background: With the widespread application of generative AI (GenAI) models, it is crucial to systematically evaluate their performance in lung adenocarcinoma histopathological assessment. This study aimed to evaluate and compare the performance of three GenAI models with visual capabilities (GPT-4o, Claude-3.5-Sonnet, and Gemini-1.5-Pro) in lung adenocarcinoma histological pattern recognition and grading, and to explore the construction of prognostic prediction models based on GenAI feature extraction. Methods: This retrospective study extracted 310 diagnostic slides from the TCGA-LUAD database for model evaluation. An additional 87 diagnostic pathology slides from local lung adenocarcinoma surgical patients were used for external validation of the prognostic model. Primary outcomes were GenAI grading accuracy and stability, measured by the area under the receiver operating characteristic curve (AUC) and intraclass correlation coefficient (ICC), respectively. Secondary outcomes included the construction and assessment of machine learning-based prognostic prediction models, utilizing features extracted by GenAI, with model performance evaluated using the Concordance index (C-index). Findings: Claude-3.5-Sonnet demonstrated the best overall performance, combining high grading accuracy (average AUC = 0.82) with moderate stability (ICC = 0.59) The optimal machine learning-based prognostic model, constructed using features extracted by Claude-3.5-Sonnet and incorporating clinical variables, showed good performance in both internal and external validation, with an average C-index of 0.72. Meta-analysis demonstrated that this prognostic model effectively stratified patients into risk groups, with the high-risk group showing significantly worse outcomes (Hazard ratio = 6.44, 95% confidence interval = 3.42-12.14). Interpretation: This study demonstrates the potential application value of GenAI models in lung adenocarcinoma histopathological assessment. Claude-3.5-Sonnet demonstrated the highest grading accuracy, and the machine learning-based prognostic model that utilized its feature extraction showed good predictive capabilities. These findings provide new research directions for AI-assisted pathological diagnosis and prognostic prediction, with the potential to improve the management of lung adenocarcinoma patients.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1016/j.lanwpc.2024.101352
- OA Status
- gold
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4407631405
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4407631405Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1016/j.lanwpc.2024.101352Digital Object Identifier
- Title
-
Evaluating generative AI models for explainable pathological feature extraction in lung adenocarcinoma: grading assessment and prognostic model constructionWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-02-01Full publication date if available
- Authors
-
Junyi Shen, Anqi Lin, Ting Wei, J. Andrew Zhang, Peng LuoList of authors in order
- Landing page
-
https://doi.org/10.1016/j.lanwpc.2024.101352Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1016/j.lanwpc.2024.101352Direct OA link when available
- Concepts
-
Grading (engineering), Pathological, Generative grammar, Generative model, Artificial intelligence, Adenocarcinoma, Computer science, Lung, Pathology, Medicine, Natural language processing, Internal medicine, Biology, Cancer, EcologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4407631405 |
|---|---|
| doi | https://doi.org/10.1016/j.lanwpc.2024.101352 |
| ids.doi | https://doi.org/10.1016/j.lanwpc.2024.101352 |
| ids.openalex | https://openalex.org/W4407631405 |
| fwci | 0.0 |
| type | article |
| title | Evaluating generative AI models for explainable pathological feature extraction in lung adenocarcinoma: grading assessment and prognostic model construction |
| biblio.issue | |
| biblio.volume | 55 |
| biblio.last_page | 101352 |
| biblio.first_page | 101352 |
| topics[0].id | https://openalex.org/T12422 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9775000214576721 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2741 |
| topics[0].subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| topics[0].display_name | Radiomics and Machine Learning in Medical Imaging |
| topics[1].id | https://openalex.org/T10862 |
| topics[1].field.id | https://openalex.org/fields/17 |
| topics[1].field.display_name | Computer Science |
| topics[1].score | 0.9440000057220459 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1702 |
| topics[1].subfield.display_name | Artificial Intelligence |
| topics[1].display_name | AI in cancer detection |
| is_xpac | False |
| apc_list.value | 3500 |
| apc_list.currency | USD |
| apc_list.value_usd | 3500 |
| apc_paid.value | 3500 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 3500 |
| concepts[0].id | https://openalex.org/C2777286243 |
| concepts[0].level | 2 |
| concepts[0].score | 0.7641485333442688 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q5591926 |
| concepts[0].display_name | Grading (engineering) |
| concepts[1].id | https://openalex.org/C207886595 |
| concepts[1].level | 2 |
| concepts[1].score | 0.5907183289527893 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1456138 |
| concepts[1].display_name | Pathological |
| concepts[2].id | https://openalex.org/C39890363 |
| concepts[2].level | 2 |
| concepts[2].score | 0.5216715335845947 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q36108 |
| concepts[2].display_name | Generative grammar |
| concepts[3].id | https://openalex.org/C167966045 |
| concepts[3].level | 3 |
| concepts[3].score | 0.4734239876270294 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q5532625 |
| concepts[3].display_name | Generative model |
| concepts[4].id | https://openalex.org/C154945302 |
| concepts[4].level | 1 |
| concepts[4].score | 0.4696938991546631 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q11660 |
| concepts[4].display_name | Artificial intelligence |
| concepts[5].id | https://openalex.org/C2781182431 |
| concepts[5].level | 3 |
| concepts[5].score | 0.46918773651123047 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q356033 |
| concepts[5].display_name | Adenocarcinoma |
| concepts[6].id | https://openalex.org/C41008148 |
| concepts[6].level | 0 |
| concepts[6].score | 0.4518525004386902 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[6].display_name | Computer science |
| concepts[7].id | https://openalex.org/C2777714996 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4139360785484314 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7886 |
| concepts[7].display_name | Lung |
| concepts[8].id | https://openalex.org/C142724271 |
| concepts[8].level | 1 |
| concepts[8].score | 0.3823889493942261 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[8].display_name | Pathology |
| concepts[9].id | https://openalex.org/C71924100 |
| concepts[9].level | 0 |
| concepts[9].score | 0.3519461750984192 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[9].display_name | Medicine |
| concepts[10].id | https://openalex.org/C204321447 |
| concepts[10].level | 1 |
| concepts[10].score | 0.3349328935146332 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q30642 |
| concepts[10].display_name | Natural language processing |
| concepts[11].id | https://openalex.org/C126322002 |
| concepts[11].level | 1 |
| concepts[11].score | 0.18392592668533325 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q11180 |
| concepts[11].display_name | Internal medicine |
| concepts[12].id | https://openalex.org/C86803240 |
| concepts[12].level | 0 |
| concepts[12].score | 0.14927664399147034 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[12].display_name | Biology |
| concepts[13].id | https://openalex.org/C121608353 |
| concepts[13].level | 2 |
| concepts[13].score | 0.0 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q12078 |
| concepts[13].display_name | Cancer |
| concepts[14].id | https://openalex.org/C18903297 |
| concepts[14].level | 1 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7150 |
| concepts[14].display_name | Ecology |
| keywords[0].id | https://openalex.org/keywords/grading |
| keywords[0].score | 0.7641485333442688 |
| keywords[0].display_name | Grading (engineering) |
| keywords[1].id | https://openalex.org/keywords/pathological |
| keywords[1].score | 0.5907183289527893 |
| keywords[1].display_name | Pathological |
| keywords[2].id | https://openalex.org/keywords/generative-grammar |
| keywords[2].score | 0.5216715335845947 |
| keywords[2].display_name | Generative grammar |
| keywords[3].id | https://openalex.org/keywords/generative-model |
| keywords[3].score | 0.4734239876270294 |
| keywords[3].display_name | Generative model |
| keywords[4].id | https://openalex.org/keywords/artificial-intelligence |
| keywords[4].score | 0.4696938991546631 |
| keywords[4].display_name | Artificial intelligence |
| keywords[5].id | https://openalex.org/keywords/adenocarcinoma |
| keywords[5].score | 0.46918773651123047 |
| keywords[5].display_name | Adenocarcinoma |
| keywords[6].id | https://openalex.org/keywords/computer-science |
| keywords[6].score | 0.4518525004386902 |
| keywords[6].display_name | Computer science |
| keywords[7].id | https://openalex.org/keywords/lung |
| keywords[7].score | 0.4139360785484314 |
| keywords[7].display_name | Lung |
| keywords[8].id | https://openalex.org/keywords/pathology |
| keywords[8].score | 0.3823889493942261 |
| keywords[8].display_name | Pathology |
| keywords[9].id | https://openalex.org/keywords/medicine |
| keywords[9].score | 0.3519461750984192 |
| keywords[9].display_name | Medicine |
| keywords[10].id | https://openalex.org/keywords/natural-language-processing |
| keywords[10].score | 0.3349328935146332 |
| keywords[10].display_name | Natural language processing |
| keywords[11].id | https://openalex.org/keywords/internal-medicine |
| keywords[11].score | 0.18392592668533325 |
| keywords[11].display_name | Internal medicine |
| keywords[12].id | https://openalex.org/keywords/biology |
| keywords[12].score | 0.14927664399147034 |
| keywords[12].display_name | Biology |
| language | en |
| locations[0].id | doi:10.1016/j.lanwpc.2024.101352 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210225048 |
| locations[0].source.issn | 2666-6065 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2666-6065 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | The Lancet Regional Health - Western Pacific |
| locations[0].source.host_organization | https://openalex.org/P4310320990 |
| locations[0].source.host_organization_name | Elsevier BV |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320990 |
| locations[0].source.host_organization_lineage_names | Elsevier BV |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | The Lancet Regional Health - Western Pacific |
| locations[0].landing_page_url | https://doi.org/10.1016/j.lanwpc.2024.101352 |
| locations[1].id | pmh:oai:doaj.org/article:5da10e828ef94e0981c0369b7be51586 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306401280 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | submittedVersion |
| locations[1].raw_type | article |
| locations[1].license_id | |
| locations[1].is_accepted | False |
| locations[1].is_published | False |
| locations[1].raw_source_name | The Lancet Regional Health. Western Pacific, Vol 55, Iss , Pp 101352- (2025) |
| locations[1].landing_page_url | https://doaj.org/article/5da10e828ef94e0981c0369b7be51586 |
| indexed_in | crossref, doaj |
| authorships[0].author.id | https://openalex.org/A5072228856 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-5859-1382 |
| authorships[0].author.display_name | Junyi Shen |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Junyi Shen |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5082582836 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-6324-0410 |
| authorships[1].author.display_name | Anqi Lin |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Anqi Lin |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5061308099 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-7840-1013 |
| authorships[2].author.display_name | Ting Wei |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Ting Wei |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5100706723 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-6102-3762 |
| authorships[3].author.display_name | J. Andrew Zhang |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Jian Zhang |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5050960828 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-8215-2045 |
| authorships[4].author.display_name | Peng Luo |
| authorships[4].author_position | last |
| authorships[4].raw_author_name | Peng Luo |
| authorships[4].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1016/j.lanwpc.2024.101352 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Evaluating generative AI models for explainable pathological feature extraction in lung adenocarcinoma: grading assessment and prognostic model construction |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12422 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9775000214576721 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2741 |
| primary_topic.subfield.display_name | Radiology, Nuclear Medicine and Imaging |
| primary_topic.display_name | Radiomics and Machine Learning in Medical Imaging |
| related_works | https://openalex.org/W4365211920, https://openalex.org/W2364892929, https://openalex.org/W3014948380, https://openalex.org/W4391584540, https://openalex.org/W4380551139, https://openalex.org/W4317695495, https://openalex.org/W4395044357, https://openalex.org/W4287117424, https://openalex.org/W4387506531, https://openalex.org/W2087346071 |
| cited_by_count | 0 |
| locations_count | 2 |
| best_oa_location.id | doi:10.1016/j.lanwpc.2024.101352 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210225048 |
| best_oa_location.source.issn | 2666-6065 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2666-6065 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | The Lancet Regional Health - Western Pacific |
| best_oa_location.source.host_organization | https://openalex.org/P4310320990 |
| best_oa_location.source.host_organization_name | Elsevier BV |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320990 |
| best_oa_location.source.host_organization_lineage_names | Elsevier BV |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | The Lancet Regional Health - Western Pacific |
| best_oa_location.landing_page_url | https://doi.org/10.1016/j.lanwpc.2024.101352 |
| primary_location.id | doi:10.1016/j.lanwpc.2024.101352 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210225048 |
| primary_location.source.issn | 2666-6065 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2666-6065 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | The Lancet Regional Health - Western Pacific |
| primary_location.source.host_organization | https://openalex.org/P4310320990 |
| primary_location.source.host_organization_name | Elsevier BV |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320990 |
| primary_location.source.host_organization_lineage_names | Elsevier BV |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | The Lancet Regional Health - Western Pacific |
| primary_location.landing_page_url | https://doi.org/10.1016/j.lanwpc.2024.101352 |
| publication_date | 2025-02-01 |
| publication_year | 2025 |
| referenced_works_count | 0 |
| abstract_inverted_index.= | 166, 172, 227, 232 |
| abstract_inverted_index.87 | 82 |
| abstract_inverted_index.AI | 7 |
| abstract_inverted_index.An | 80 |
| abstract_inverted_index.an | 200 |
| abstract_inverted_index.by | 110, 142, 184 |
| abstract_inverted_index.in | 18, 43, 193, 245 |
| abstract_inverted_index.is | 11 |
| abstract_inverted_index.it | 10 |
| abstract_inverted_index.of | 5, 32, 56, 97, 133, 203, 242, 291 |
| abstract_inverted_index.on | 61 |
| abstract_inverted_index.to | 13, 26, 52, 287 |
| abstract_inverted_index.310 | 70 |
| abstract_inverted_index.95% | 229 |
| abstract_inverted_index.AUC | 165 |
| abstract_inverted_index.The | 174 |
| abstract_inverted_index.and | 28, 41, 49, 51, 107, 120, 131, 186, 196, 256, 281 |
| abstract_inverted_index.for | 77, 94, 277 |
| abstract_inverted_index.its | 264 |
| abstract_inverted_index.new | 274 |
| abstract_inverted_index.the | 2, 30, 54, 74, 98, 111, 114, 129, 149, 156, 218, 238, 252, 257, 285, 289 |
| abstract_inverted_index.(ICC | 171 |
| abstract_inverted_index.This | 23, 66, 235 |
| abstract_inverted_index.With | 1 |
| abstract_inverted_index.area | 112 |
| abstract_inverted_index.best | 157 |
| abstract_inverted_index.both | 194 |
| abstract_inverted_index.from | 73, 86 |
| abstract_inverted_index.good | 191, 268 |
| abstract_inverted_index.high | 161 |
| abstract_inverted_index.into | 214 |
| abstract_inverted_index.lung | 19, 44, 88, 246, 292 |
| abstract_inverted_index.risk | 215 |
| abstract_inverted_index.that | 207, 262 |
| abstract_inverted_index.this | 208 |
| abstract_inverted_index.used | 93 |
| abstract_inverted_index.were | 92, 103 |
| abstract_inverted_index.with | 36, 144, 168, 199, 217, 284 |
| abstract_inverted_index.(AUC) | 119 |
| abstract_inverted_index.0.59) | 173 |
| abstract_inverted_index.0.72. | 204 |
| abstract_inverted_index.0.82) | 167 |
| abstract_inverted_index.6.44, | 228 |
| abstract_inverted_index.GenAI | 34, 62, 104, 243 |
| abstract_inverted_index.These | 271 |
| abstract_inverted_index.aimed | 25 |
| abstract_inverted_index.based | 60 |
| abstract_inverted_index.curve | 118 |
| abstract_inverted_index.group | 220 |
| abstract_inverted_index.index | 151 |
| abstract_inverted_index.local | 87 |
| abstract_inverted_index.model | 78, 145, 210, 261 |
| abstract_inverted_index.ratio | 226 |
| abstract_inverted_index.study | 24, 68, 236 |
| abstract_inverted_index.their | 16 |
| abstract_inverted_index.three | 33 |
| abstract_inverted_index.under | 113 |
| abstract_inverted_index.using | 148, 181 |
| abstract_inverted_index.value | 241 |
| abstract_inverted_index.worse | 223 |
| abstract_inverted_index.(ICC), | 124 |
| abstract_inverted_index.GenAI, | 143 |
| abstract_inverted_index.model, | 179 |
| abstract_inverted_index.model. | 100 |
| abstract_inverted_index.models | 35, 59, 244 |
| abstract_inverted_index.showed | 190, 267 |
| abstract_inverted_index.slides | 72, 85 |
| abstract_inverted_index.visual | 37 |
| abstract_inverted_index.(GenAI) | 8 |
| abstract_inverted_index.(Hazard | 225 |
| abstract_inverted_index.C-index | 202 |
| abstract_inverted_index.Primary | 101 |
| abstract_inverted_index.average | 201 |
| abstract_inverted_index.compare | 29 |
| abstract_inverted_index.crucial | 12 |
| abstract_inverted_index.explore | 53 |
| abstract_inverted_index.feature | 63, 265 |
| abstract_inverted_index.grading | 105, 162, 254 |
| abstract_inverted_index.groups, | 216 |
| abstract_inverted_index.highest | 253 |
| abstract_inverted_index.improve | 288 |
| abstract_inverted_index.machine | 134, 176, 258 |
| abstract_inverted_index.models, | 9, 138 |
| abstract_inverted_index.optimal | 175 |
| abstract_inverted_index.overall | 158 |
| abstract_inverted_index.pattern | 47 |
| abstract_inverted_index.provide | 273 |
| abstract_inverted_index.showing | 221 |
| abstract_inverted_index.(GPT-4o, | 39 |
| abstract_inverted_index.(average | 164 |
| abstract_inverted_index.Methods: | 65 |
| abstract_inverted_index.accuracy | 106, 163 |
| abstract_inverted_index.clinical | 188 |
| abstract_inverted_index.database | 76 |
| abstract_inverted_index.evaluate | 15, 27 |
| abstract_inverted_index.external | 95, 197 |
| abstract_inverted_index.features | 140, 182 |
| abstract_inverted_index.findings | 272 |
| abstract_inverted_index.grading, | 50 |
| abstract_inverted_index.included | 128 |
| abstract_inverted_index.internal | 195 |
| abstract_inverted_index.interval | 231 |
| abstract_inverted_index.measured | 109 |
| abstract_inverted_index.moderate | 169 |
| abstract_inverted_index.outcomes | 102, 127, 224 |
| abstract_inverted_index.patients | 91, 213 |
| abstract_inverted_index.receiver | 115 |
| abstract_inverted_index.research | 275 |
| abstract_inverted_index.surgical | 90 |
| abstract_inverted_index.utilized | 263 |
| abstract_inverted_index.Findings: | 153 |
| abstract_inverted_index.Secondary | 126 |
| abstract_inverted_index.TCGA-LUAD | 75 |
| abstract_inverted_index.accuracy, | 255 |
| abstract_inverted_index.combining | 160 |
| abstract_inverted_index.diagnosis | 280 |
| abstract_inverted_index.evaluated | 147 |
| abstract_inverted_index.extracted | 69, 141, 183 |
| abstract_inverted_index.high-risk | 219 |
| abstract_inverted_index.operating | 116 |
| abstract_inverted_index.pathology | 84 |
| abstract_inverted_index.patients. | 294 |
| abstract_inverted_index.potential | 239, 286 |
| abstract_inverted_index.stability | 170 |
| abstract_inverted_index.utilizing | 139 |
| abstract_inverted_index.(C-index). | 152 |
| abstract_inverted_index.additional | 81 |
| abstract_inverted_index.assessment | 132 |
| abstract_inverted_index.confidence | 230 |
| abstract_inverted_index.diagnostic | 71, 83 |
| abstract_inverted_index.directions | 276 |
| abstract_inverted_index.extraction | 266 |
| abstract_inverted_index.generative | 6 |
| abstract_inverted_index.intraclass | 121 |
| abstract_inverted_index.management | 290 |
| abstract_inverted_index.prediction | 58, 137 |
| abstract_inverted_index.predictive | 269 |
| abstract_inverted_index.prognostic | 57, 99, 136, 178, 209, 260, 282 |
| abstract_inverted_index.stability, | 108 |
| abstract_inverted_index.stratified | 212 |
| abstract_inverted_index.validation | 96 |
| abstract_inverted_index.variables, | 189 |
| abstract_inverted_index.widespread | 3 |
| abstract_inverted_index.AI-assisted | 278 |
| abstract_inverted_index.Background: | 0 |
| abstract_inverted_index.Concordance | 150 |
| abstract_inverted_index.application | 4, 240 |
| abstract_inverted_index.assessment. | 22, 249 |
| abstract_inverted_index.coefficient | 123 |
| abstract_inverted_index.constructed | 180 |
| abstract_inverted_index.correlation | 122 |
| abstract_inverted_index.effectively | 211 |
| abstract_inverted_index.evaluation. | 79 |
| abstract_inverted_index.extraction. | 64 |
| abstract_inverted_index.performance | 17, 31, 146, 192 |
| abstract_inverted_index.prediction, | 283 |
| abstract_inverted_index.recognition | 48 |
| abstract_inverted_index.validation, | 198 |
| abstract_inverted_index.3.42-12.14). | 233 |
| abstract_inverted_index.capabilities | 38 |
| abstract_inverted_index.construction | 55, 130 |
| abstract_inverted_index.demonstrated | 155, 206, 251 |
| abstract_inverted_index.demonstrates | 237 |
| abstract_inverted_index.histological | 46 |
| abstract_inverted_index.pathological | 279 |
| abstract_inverted_index.performance, | 159 |
| abstract_inverted_index.Meta-analysis | 205 |
| abstract_inverted_index.capabilities. | 270 |
| abstract_inverted_index.incorporating | 187 |
| abstract_inverted_index.respectively. | 125 |
| abstract_inverted_index.retrospective | 67 |
| abstract_inverted_index.significantly | 222 |
| abstract_inverted_index.adenocarcinoma | 20, 45, 89, 247, 293 |
| abstract_inverted_index.characteristic | 117 |
| abstract_inverted_index.learning-based | 135, 177, 259 |
| abstract_inverted_index.systematically | 14 |
| abstract_inverted_index.Gemini-1.5-Pro) | 42 |
| abstract_inverted_index.Interpretation: | 234 |
| abstract_inverted_index.Claude-3.5-Sonnet | 154, 185, 250 |
| abstract_inverted_index.histopathological | 21, 248 |
| abstract_inverted_index.Claude-3.5-Sonnet, | 40 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 5 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/2 |
| sustainable_development_goals[0].score | 0.5199999809265137 |
| sustainable_development_goals[0].display_name | Zero hunger |
| citation_normalized_percentile.value | 0.06454101 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |