Evaluation of a Two-Stage Statistical Learning Design for Genome-Wide Studies Article Swipe
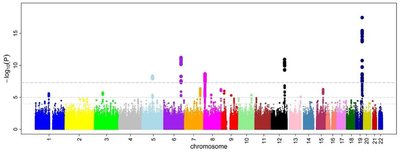
Twin and family studies show that many common traits and disorders are highly heritable, but genome-wide association studies (GWAS) have been largely unable to identify specific single nucleotide polymorphisms (SNPs) explaining this heritability at the genetic level. Recent work suggests statistical learning methods like gradient boosting (GBM) may be a viable alternative to conventional methods, especially after adjustments for the structure of SNP data. The current research evaluates a two-stage research design for GWAS. GBM is used as a first stage variable selection screen to substantially reduce the dimensionality of SNP data while maintaining sensitivity to additive, nonlinear, and interaction effects, allowing hypothesis testing with a reduced multiple testing burden in the second stage analysis. Thorough simulations shows the proposed two-stage design can substantially improve power to detect effect SNPs in a wide range of conditions. The limitations and potential improvements to this design are explored.
Related Topics
- Type
- article
- Language
- en
- OA Status
- green
- Related Works
- 20
- OpenAlex ID
- https://openalex.org/W2592522449
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W2592522449Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.7274/p2676t07f3dDigital Object Identifier
- Title
-
Evaluation of a Two-Stage Statistical Learning Design for Genome-Wide StudiesWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2022Year of publication
- Publication date
-
2022-09-15Full publication date if available
- Authors
-
Raymond K. WaltersList of authors in order
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- Concepts
-
Genome-wide association study, Single-nucleotide polymorphism, Genetic association, Statistical power, Heritability, SNP, Feature selection, Computer science, Computational biology, Biology, Genetics, Machine learning, Statistics, Mathematics, Genotype, GeneTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- Related works (count)
-
20Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W2592522449 |
|---|---|
| doi | https://doi.org/10.7274/p2676t07f3d |
| ids.doi | https://doi.org/10.7274/p2676t07f3d |
| ids.mag | 2592522449 |
| ids.openalex | https://openalex.org/W2592522449 |
| fwci | 0.0 |
| type | article |
| title | Evaluation of a Two-Stage Statistical Learning Design for Genome-Wide Studies |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10594 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9961000084877014 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Genetic and phenotypic traits in livestock |
| topics[1].id | https://openalex.org/T10885 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9945999979972839 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | Gene expression and cancer classification |
| topics[2].id | https://openalex.org/T11468 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9919999837875366 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1311 |
| topics[2].subfield.display_name | Genetics |
| topics[2].display_name | Genetic Mapping and Diversity in Plants and Animals |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C106208931 |
| concepts[0].level | 5 |
| concepts[0].score | 0.7264155149459839 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[0].display_name | Genome-wide association study |
| concepts[1].id | https://openalex.org/C153209595 |
| concepts[1].level | 4 |
| concepts[1].score | 0.6060545444488525 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[1].display_name | Single-nucleotide polymorphism |
| concepts[2].id | https://openalex.org/C186413461 |
| concepts[2].level | 5 |
| concepts[2].score | 0.5873688459396362 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[2].display_name | Genetic association |
| concepts[3].id | https://openalex.org/C96608239 |
| concepts[3].level | 2 |
| concepts[3].score | 0.49810075759887695 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q1199823 |
| concepts[3].display_name | Statistical power |
| concepts[4].id | https://openalex.org/C161890455 |
| concepts[4].level | 2 |
| concepts[4].score | 0.4736163318157196 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1503548 |
| concepts[4].display_name | Heritability |
| concepts[5].id | https://openalex.org/C139275648 |
| concepts[5].level | 5 |
| concepts[5].score | 0.4553409814834595 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q17134011 |
| concepts[5].display_name | SNP |
| concepts[6].id | https://openalex.org/C148483581 |
| concepts[6].level | 2 |
| concepts[6].score | 0.4482571482658386 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q446488 |
| concepts[6].display_name | Feature selection |
| concepts[7].id | https://openalex.org/C41008148 |
| concepts[7].level | 0 |
| concepts[7].score | 0.4308801293373108 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[7].display_name | Computer science |
| concepts[8].id | https://openalex.org/C70721500 |
| concepts[8].level | 1 |
| concepts[8].score | 0.38060763478279114 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[8].display_name | Computational biology |
| concepts[9].id | https://openalex.org/C86803240 |
| concepts[9].level | 0 |
| concepts[9].score | 0.3792261779308319 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[9].display_name | Biology |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.320917546749115 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C119857082 |
| concepts[11].level | 1 |
| concepts[11].score | 0.3035435080528259 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q2539 |
| concepts[11].display_name | Machine learning |
| concepts[12].id | https://openalex.org/C105795698 |
| concepts[12].level | 1 |
| concepts[12].score | 0.24409529566764832 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q12483 |
| concepts[12].display_name | Statistics |
| concepts[13].id | https://openalex.org/C33923547 |
| concepts[13].level | 0 |
| concepts[13].score | 0.1773744821548462 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q395 |
| concepts[13].display_name | Mathematics |
| concepts[14].id | https://openalex.org/C135763542 |
| concepts[14].level | 3 |
| concepts[14].score | 0.08720442652702332 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[14].display_name | Genotype |
| concepts[15].id | https://openalex.org/C104317684 |
| concepts[15].level | 2 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[15].display_name | Gene |
| keywords[0].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[0].score | 0.7264155149459839 |
| keywords[0].display_name | Genome-wide association study |
| keywords[1].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[1].score | 0.6060545444488525 |
| keywords[1].display_name | Single-nucleotide polymorphism |
| keywords[2].id | https://openalex.org/keywords/genetic-association |
| keywords[2].score | 0.5873688459396362 |
| keywords[2].display_name | Genetic association |
| keywords[3].id | https://openalex.org/keywords/statistical-power |
| keywords[3].score | 0.49810075759887695 |
| keywords[3].display_name | Statistical power |
| keywords[4].id | https://openalex.org/keywords/heritability |
| keywords[4].score | 0.4736163318157196 |
| keywords[4].display_name | Heritability |
| keywords[5].id | https://openalex.org/keywords/snp |
| keywords[5].score | 0.4553409814834595 |
| keywords[5].display_name | SNP |
| keywords[6].id | https://openalex.org/keywords/feature-selection |
| keywords[6].score | 0.4482571482658386 |
| keywords[6].display_name | Feature selection |
| keywords[7].id | https://openalex.org/keywords/computer-science |
| keywords[7].score | 0.4308801293373108 |
| keywords[7].display_name | Computer science |
| keywords[8].id | https://openalex.org/keywords/computational-biology |
| keywords[8].score | 0.38060763478279114 |
| keywords[8].display_name | Computational biology |
| keywords[9].id | https://openalex.org/keywords/biology |
| keywords[9].score | 0.3792261779308319 |
| keywords[9].display_name | Biology |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.320917546749115 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/machine-learning |
| keywords[11].score | 0.3035435080528259 |
| keywords[11].display_name | Machine learning |
| keywords[12].id | https://openalex.org/keywords/statistics |
| keywords[12].score | 0.24409529566764832 |
| keywords[12].display_name | Statistics |
| keywords[13].id | https://openalex.org/keywords/mathematics |
| keywords[13].score | 0.1773744821548462 |
| keywords[13].display_name | Mathematics |
| keywords[14].id | https://openalex.org/keywords/genotype |
| keywords[14].score | 0.08720442652702332 |
| keywords[14].display_name | Genotype |
| language | en |
| locations[0].id | pmh:oai:figshare.com:article/24826755 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306400572 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| locations[0].source.host_organization | https://openalex.org/I196829312 |
| locations[0].source.host_organization_name | La Trobe University |
| locations[0].source.host_organization_lineage | https://openalex.org/I196829312 |
| locations[0].license | |
| locations[0].pdf_url | |
| locations[0].version | submittedVersion |
| locations[0].raw_type | Text |
| locations[0].license_id | |
| locations[0].is_accepted | False |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | |
| locations[1].id | doi:10.7274/p2676t07f3d |
| locations[1].is_oa | True |
| locations[1].source.id | https://openalex.org/S7407053147 |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | University of Notre Dame |
| locations[1].source.host_organization | |
| locations[1].source.host_organization_name | |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | |
| locations[1].raw_type | article-journal |
| locations[1].license_id | |
| locations[1].is_accepted | False |
| locations[1].is_published | |
| locations[1].raw_source_name | |
| locations[1].landing_page_url | https://doi.org/10.7274/p2676t07f3d |
| locations[2].id | mag:2592522449 |
| locations[2].is_oa | False |
| locations[2].source | |
| locations[2].license | |
| locations[2].pdf_url | |
| locations[2].version | |
| locations[2].raw_type | |
| locations[2].license_id | |
| locations[2].is_accepted | False |
| locations[2].is_published | |
| locations[2].raw_source_name | |
| locations[2].landing_page_url | https://oatd.org/oatd/record?record=oai\:notredame\:p2676t07f3d |
| indexed_in | datacite |
| authorships[0].author.id | https://openalex.org/A5091572489 |
| authorships[0].author.orcid | https://orcid.org/0000-0001-8422-6530 |
| authorships[0].author.display_name | Raymond K. Walters |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Raymond Kenney Walters |
| authorships[0].is_corresponding | True |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Evaluation of a Two-Stage Statistical Learning Design for Genome-Wide Studies |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T06:51:31.235846 |
| primary_topic.id | https://openalex.org/T10594 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9961000084877014 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Genetic and phenotypic traits in livestock |
| related_works | https://openalex.org/W3092278780, https://openalex.org/W2085037019, https://openalex.org/W1642438486, https://openalex.org/W2789896172, https://openalex.org/W3016610742, https://openalex.org/W262550870, https://openalex.org/W2951415774, https://openalex.org/W3201164482, https://openalex.org/W2596895167, https://openalex.org/W1903006068, https://openalex.org/W1502118703, https://openalex.org/W2901004011, https://openalex.org/W2150990910, https://openalex.org/W2000113303, https://openalex.org/W2971418588, https://openalex.org/W2565205739, https://openalex.org/W2997786719, https://openalex.org/W2799207035, https://openalex.org/W2747134847, https://openalex.org/W1832916950 |
| cited_by_count | 0 |
| locations_count | 3 |
| best_oa_location.id | pmh:oai:figshare.com:article/24826755 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306400572 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| best_oa_location.source.host_organization | https://openalex.org/I196829312 |
| best_oa_location.source.host_organization_name | La Trobe University |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I196829312 |
| best_oa_location.license | |
| best_oa_location.pdf_url | |
| best_oa_location.version | submittedVersion |
| best_oa_location.raw_type | Text |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | False |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | |
| primary_location.id | pmh:oai:figshare.com:article/24826755 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306400572 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | OPAL (Open@LaTrobe) (La Trobe University) |
| primary_location.source.host_organization | https://openalex.org/I196829312 |
| primary_location.source.host_organization_name | La Trobe University |
| primary_location.source.host_organization_lineage | https://openalex.org/I196829312 |
| primary_location.license | |
| primary_location.pdf_url | |
| primary_location.version | submittedVersion |
| primary_location.raw_type | Text |
| primary_location.license_id | |
| primary_location.is_accepted | False |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | |
| publication_date | 2022-09-15 |
| publication_year | 2022 |
| referenced_works_count | 0 |
| abstract_inverted_index.a | 49, 68, 78, 105, 131 |
| abstract_inverted_index.as | 77 |
| abstract_inverted_index.at | 33 |
| abstract_inverted_index.be | 48 |
| abstract_inverted_index.in | 110, 130 |
| abstract_inverted_index.is | 75 |
| abstract_inverted_index.of | 61, 89, 134 |
| abstract_inverted_index.to | 23, 52, 84, 95, 126, 141 |
| abstract_inverted_index.GBM | 74 |
| abstract_inverted_index.SNP | 62, 90 |
| abstract_inverted_index.The | 64, 136 |
| abstract_inverted_index.and | 1, 9, 98, 138 |
| abstract_inverted_index.are | 11, 144 |
| abstract_inverted_index.but | 14 |
| abstract_inverted_index.can | 122 |
| abstract_inverted_index.for | 58, 72 |
| abstract_inverted_index.may | 47 |
| abstract_inverted_index.the | 34, 59, 87, 111, 118 |
| abstract_inverted_index.SNPs | 129 |
| abstract_inverted_index.Twin | 0 |
| abstract_inverted_index.been | 20 |
| abstract_inverted_index.data | 91 |
| abstract_inverted_index.have | 19 |
| abstract_inverted_index.like | 43 |
| abstract_inverted_index.many | 6 |
| abstract_inverted_index.show | 4 |
| abstract_inverted_index.that | 5 |
| abstract_inverted_index.this | 31, 142 |
| abstract_inverted_index.used | 76 |
| abstract_inverted_index.wide | 132 |
| abstract_inverted_index.with | 104 |
| abstract_inverted_index.work | 38 |
| abstract_inverted_index.(GBM) | 46 |
| abstract_inverted_index.GWAS. | 73 |
| abstract_inverted_index.after | 56 |
| abstract_inverted_index.data. | 63 |
| abstract_inverted_index.first | 79 |
| abstract_inverted_index.power | 125 |
| abstract_inverted_index.range | 133 |
| abstract_inverted_index.shows | 117 |
| abstract_inverted_index.stage | 80, 113 |
| abstract_inverted_index.while | 92 |
| abstract_inverted_index.(GWAS) | 18 |
| abstract_inverted_index.(SNPs) | 29 |
| abstract_inverted_index.Recent | 37 |
| abstract_inverted_index.burden | 109 |
| abstract_inverted_index.common | 7 |
| abstract_inverted_index.design | 71, 121, 143 |
| abstract_inverted_index.detect | 127 |
| abstract_inverted_index.effect | 128 |
| abstract_inverted_index.family | 2 |
| abstract_inverted_index.highly | 12 |
| abstract_inverted_index.level. | 36 |
| abstract_inverted_index.reduce | 86 |
| abstract_inverted_index.screen | 83 |
| abstract_inverted_index.second | 112 |
| abstract_inverted_index.single | 26 |
| abstract_inverted_index.traits | 8 |
| abstract_inverted_index.unable | 22 |
| abstract_inverted_index.viable | 50 |
| abstract_inverted_index.current | 65 |
| abstract_inverted_index.genetic | 35 |
| abstract_inverted_index.improve | 124 |
| abstract_inverted_index.largely | 21 |
| abstract_inverted_index.methods | 42 |
| abstract_inverted_index.reduced | 106 |
| abstract_inverted_index.studies | 3, 17 |
| abstract_inverted_index.testing | 103, 108 |
| abstract_inverted_index.Thorough | 115 |
| abstract_inverted_index.allowing | 101 |
| abstract_inverted_index.boosting | 45 |
| abstract_inverted_index.effects, | 100 |
| abstract_inverted_index.gradient | 44 |
| abstract_inverted_index.identify | 24 |
| abstract_inverted_index.learning | 41 |
| abstract_inverted_index.methods, | 54 |
| abstract_inverted_index.multiple | 107 |
| abstract_inverted_index.proposed | 119 |
| abstract_inverted_index.research | 66, 70 |
| abstract_inverted_index.specific | 25 |
| abstract_inverted_index.suggests | 39 |
| abstract_inverted_index.variable | 81 |
| abstract_inverted_index.additive, | 96 |
| abstract_inverted_index.analysis. | 114 |
| abstract_inverted_index.disorders | 10 |
| abstract_inverted_index.evaluates | 67 |
| abstract_inverted_index.explored. | 145 |
| abstract_inverted_index.potential | 139 |
| abstract_inverted_index.selection | 82 |
| abstract_inverted_index.structure | 60 |
| abstract_inverted_index.two-stage | 69, 120 |
| abstract_inverted_index.especially | 55 |
| abstract_inverted_index.explaining | 30 |
| abstract_inverted_index.heritable, | 13 |
| abstract_inverted_index.hypothesis | 102 |
| abstract_inverted_index.nonlinear, | 97 |
| abstract_inverted_index.nucleotide | 27 |
| abstract_inverted_index.adjustments | 57 |
| abstract_inverted_index.alternative | 51 |
| abstract_inverted_index.association | 16 |
| abstract_inverted_index.conditions. | 135 |
| abstract_inverted_index.genome-wide | 15 |
| abstract_inverted_index.interaction | 99 |
| abstract_inverted_index.limitations | 137 |
| abstract_inverted_index.maintaining | 93 |
| abstract_inverted_index.sensitivity | 94 |
| abstract_inverted_index.simulations | 116 |
| abstract_inverted_index.statistical | 40 |
| abstract_inverted_index.conventional | 53 |
| abstract_inverted_index.heritability | 32 |
| abstract_inverted_index.improvements | 140 |
| abstract_inverted_index.polymorphisms | 28 |
| abstract_inverted_index.substantially | 85, 123 |
| abstract_inverted_index.dimensionality | 88 |
| cited_by_percentile_year | |
| corresponding_author_ids | https://openalex.org/A5091572489 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 1 |
| citation_normalized_percentile.value | 0.00079785 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |