Four Loss of Function Pathogenic Variants in ARHGAP29 in Non-Syndromic Cleft Lip and Palate Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.21203/rs.3.rs-4557690/v1
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.21203/rs.3.rs-4557690/v1
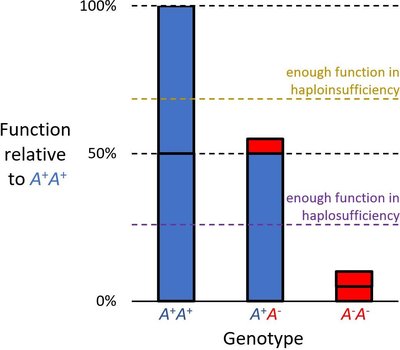
The pathophysiological basis of non-syndromic cleft lip and/or palate (NsCL/P) is still largely unclear. However, exome sequencing (ES) has allowed to associate several genes with NsCL/P, often with reduced penetrance. Among these genes, the Rho GTPase activating protein 29 (ARHGAP29) has been previously implicated in 7 families with NsCL/P. We investigated a cohort of 224 NsCLPs for which no genetic mutation had been identified by diagnostic testing. We used ES and bioinformatic variant filtering and identified four novel likely pathogenic/pathologic variants in ARHGAP29 in four multiplex families. One was a missense variant leading to the substitution of the first methionine with threonine, two were heterozygous frameshift variants leading to a premature termination codon, and the last one was a nonsense variant. All variants were predicted to result in loss of function, either through NMD-mediated mRNA decay, or by truncated ARHGAP29, or by non-translation or abnormal N-terminal initiation of translation of ARHGAP29. The truncated ARHGAP29 proteins would lack the important RhoGAP domain. The variants were not present in the control population databases, and the loss of intolerance score (pLI) of ARHGAP29 is 1.0, suggesting that ARHGAP29 haploinsufficiency is not tolerated. Phenotypes ranged from microform CL to complete bilateral CLP, with one unaffected mutation carriers. These results extend the mutational spectrum of ARHGAP29 and show that it is an important gene underlying variable NsCL/P phenotypes. ARHGAP29 should be included in diagnostic genetic testing for NsCL/P, especially familial cases, as it may be mutated in ⁓2% of patients with high penetrance (88%).
Related Topics
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.21203/rs.3.rs-4557690/v1
- OA Status
- gold
- References
- 41
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4400640442
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4400640442Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.21203/rs.3.rs-4557690/v1Digital Object Identifier
- Title
-
Four Loss of Function Pathogenic Variants in ARHGAP29 in Non-Syndromic Cleft Lip and PalateWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2024Year of publication
- Publication date
-
2024-07-15Full publication date if available
- Authors
-
Miikka Vikkula, Peyman Ranji, Eleonore Pairet, Raphaël Helaers, Bénédicte Bayet, Alexander Gerdom, Vera Lúcia Gil‐da‐Silva‐Lopes, Nicole RevençuList of authors in order
- Landing page
-
https://doi.org/10.21203/rs.3.rs-4557690/v1Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.21203/rs.3.rs-4557690/v1Direct OA link when available
- Concepts
-
Genetics, Frameshift mutation, Biology, Missense mutation, Haploinsufficiency, Gene, Mutation, PhenotypeTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
41Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4400640442 |
|---|---|
| doi | https://doi.org/10.21203/rs.3.rs-4557690/v1 |
| ids.doi | https://doi.org/10.21203/rs.3.rs-4557690/v1 |
| ids.openalex | https://openalex.org/W4400640442 |
| fwci | |
| type | preprint |
| title | Four Loss of Function Pathogenic Variants in ARHGAP29 in Non-Syndromic Cleft Lip and Palate |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T11374 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 0.9997000098228455 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1311 |
| topics[0].subfield.display_name | Genetics |
| topics[0].display_name | Cleft Lip and Palate Research |
| topics[1].id | https://openalex.org/T11679 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9940999746322632 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Craniofacial Disorders and Treatments |
| topics[2].id | https://openalex.org/T11919 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9919999837875366 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | dental development and anomalies |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C54355233 |
| concepts[0].level | 1 |
| concepts[0].score | 0.7313161492347717 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[0].display_name | Genetics |
| concepts[1].id | https://openalex.org/C29906990 |
| concepts[1].level | 4 |
| concepts[1].score | 0.7100766897201538 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q1440688 |
| concepts[1].display_name | Frameshift mutation |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.5888484716415405 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C75563809 |
| concepts[3].level | 4 |
| concepts[3].score | 0.450824499130249 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q2656896 |
| concepts[3].display_name | Missense mutation |
| concepts[4].id | https://openalex.org/C68838962 |
| concepts[4].level | 4 |
| concepts[4].score | 0.42952680587768555 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q852654 |
| concepts[4].display_name | Haploinsufficiency |
| concepts[5].id | https://openalex.org/C104317684 |
| concepts[5].level | 2 |
| concepts[5].score | 0.383818119764328 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[5].display_name | Gene |
| concepts[6].id | https://openalex.org/C501734568 |
| concepts[6].level | 3 |
| concepts[6].score | 0.36570286750793457 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q42918 |
| concepts[6].display_name | Mutation |
| concepts[7].id | https://openalex.org/C127716648 |
| concepts[7].level | 3 |
| concepts[7].score | 0.3251868784427643 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q104053 |
| concepts[7].display_name | Phenotype |
| keywords[0].id | https://openalex.org/keywords/genetics |
| keywords[0].score | 0.7313161492347717 |
| keywords[0].display_name | Genetics |
| keywords[1].id | https://openalex.org/keywords/frameshift-mutation |
| keywords[1].score | 0.7100766897201538 |
| keywords[1].display_name | Frameshift mutation |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.5888484716415405 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/missense-mutation |
| keywords[3].score | 0.450824499130249 |
| keywords[3].display_name | Missense mutation |
| keywords[4].id | https://openalex.org/keywords/haploinsufficiency |
| keywords[4].score | 0.42952680587768555 |
| keywords[4].display_name | Haploinsufficiency |
| keywords[5].id | https://openalex.org/keywords/gene |
| keywords[5].score | 0.383818119764328 |
| keywords[5].display_name | Gene |
| keywords[6].id | https://openalex.org/keywords/mutation |
| keywords[6].score | 0.36570286750793457 |
| keywords[6].display_name | Mutation |
| keywords[7].id | https://openalex.org/keywords/phenotype |
| keywords[7].score | 0.3251868784427643 |
| keywords[7].display_name | Phenotype |
| language | en |
| locations[0].id | doi:10.21203/rs.3.rs-4557690/v1 |
| locations[0].is_oa | True |
| locations[0].source | |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.21203/rs.3.rs-4557690/v1 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5091880870 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-6236-338X |
| authorships[0].author.display_name | Miikka Vikkula |
| authorships[0].countries | BE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I4210114657 |
| authorships[0].affiliations[0].raw_affiliation_string | de Duve Institute, University of Louvain |
| authorships[0].institutions[0].id | https://openalex.org/I4210114657 |
| authorships[0].institutions[0].ror | https://ror.org/022em3k58 |
| authorships[0].institutions[0].type | facility |
| authorships[0].institutions[0].lineage | https://openalex.org/I4210114657 |
| authorships[0].institutions[0].country_code | BE |
| authorships[0].institutions[0].display_name | de Duve Institute |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Miikka Vikkula |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | de Duve Institute, University of Louvain |
| authorships[1].author.id | https://openalex.org/A5104541494 |
| authorships[1].author.orcid | https://orcid.org/0000-0002-8842-1592 |
| authorships[1].author.display_name | Peyman Ranji |
| authorships[1].countries | BE |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I4210114657 |
| authorships[1].affiliations[0].raw_affiliation_string | de Duve Institute, University of Louvain |
| authorships[1].institutions[0].id | https://openalex.org/I4210114657 |
| authorships[1].institutions[0].ror | https://ror.org/022em3k58 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I4210114657 |
| authorships[1].institutions[0].country_code | BE |
| authorships[1].institutions[0].display_name | de Duve Institute |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Peyman Ranji |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | de Duve Institute, University of Louvain |
| authorships[2].author.id | https://openalex.org/A5078522757 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-0929-9101 |
| authorships[2].author.display_name | Eleonore Pairet |
| authorships[2].countries | BE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I4210114657, https://openalex.org/I95674353 |
| authorships[2].affiliations[0].raw_affiliation_string | de Duve Institute, Université Catholique de Louvain, Brussels, Belgium. |
| authorships[2].institutions[0].id | https://openalex.org/I95674353 |
| authorships[2].institutions[0].ror | https://ror.org/02495e989 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I95674353 |
| authorships[2].institutions[0].country_code | BE |
| authorships[2].institutions[0].display_name | UCLouvain |
| authorships[2].institutions[1].id | https://openalex.org/I4210114657 |
| authorships[2].institutions[1].ror | https://ror.org/022em3k58 |
| authorships[2].institutions[1].type | facility |
| authorships[2].institutions[1].lineage | https://openalex.org/I4210114657 |
| authorships[2].institutions[1].country_code | BE |
| authorships[2].institutions[1].display_name | de Duve Institute |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Eleonore Pairet |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | de Duve Institute, Université Catholique de Louvain, Brussels, Belgium. |
| authorships[3].author.id | https://openalex.org/A5047781460 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-7046-7867 |
| authorships[3].author.display_name | Raphaël Helaers |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Raphael Helaers |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5008469444 |
| authorships[4].author.orcid | |
| authorships[4].author.display_name | Bénédicte Bayet |
| authorships[4].countries | BE |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I1315288073 |
| authorships[4].affiliations[0].raw_affiliation_string | Cliniques universitaires Saint-Luc, University of Louvain |
| authorships[4].institutions[0].id | https://openalex.org/I1315288073 |
| authorships[4].institutions[0].ror | https://ror.org/03s4khd80 |
| authorships[4].institutions[0].type | healthcare |
| authorships[4].institutions[0].lineage | https://openalex.org/I1315288073 |
| authorships[4].institutions[0].country_code | BE |
| authorships[4].institutions[0].display_name | Cliniques Universitaires Saint-Luc |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Bénédicte Bayet |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Cliniques universitaires Saint-Luc, University of Louvain |
| authorships[5].author.id | https://openalex.org/A5005361599 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-7006-1296 |
| authorships[5].author.display_name | Alexander Gerdom |
| authorships[5].countries | BE |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I1315288073 |
| authorships[5].affiliations[0].raw_affiliation_string | Cliniques universitaires Saint-Luc, University of Louvain |
| authorships[5].institutions[0].id | https://openalex.org/I1315288073 |
| authorships[5].institutions[0].ror | https://ror.org/03s4khd80 |
| authorships[5].institutions[0].type | healthcare |
| authorships[5].institutions[0].lineage | https://openalex.org/I1315288073 |
| authorships[5].institutions[0].country_code | BE |
| authorships[5].institutions[0].display_name | Cliniques Universitaires Saint-Luc |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Alexander Gerdom |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Cliniques universitaires Saint-Luc, University of Louvain |
| authorships[6].author.id | https://openalex.org/A5037480487 |
| authorships[6].author.orcid | https://orcid.org/0000-0003-1288-0554 |
| authorships[6].author.display_name | Vera Lúcia Gil‐da‐Silva‐Lopes |
| authorships[6].countries | BR |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I181391015 |
| authorships[6].affiliations[0].raw_affiliation_string | UNICAMP: Universidade Estadual de Campinas |
| authorships[6].institutions[0].id | https://openalex.org/I181391015 |
| authorships[6].institutions[0].ror | https://ror.org/04wffgt70 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I181391015 |
| authorships[6].institutions[0].country_code | BR |
| authorships[6].institutions[0].display_name | Universidade Estadual de Campinas (UNICAMP) |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Vera Lucia Gil da Silva Lopes |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | UNICAMP: Universidade Estadual de Campinas |
| authorships[7].author.id | https://openalex.org/A5052857699 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-7120-4903 |
| authorships[7].author.display_name | Nicole Revençu |
| authorships[7].countries | BE |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I1315288073 |
| authorships[7].affiliations[0].raw_affiliation_string | Cliniques universitaires Saint-Luc, University of Louvain |
| authorships[7].institutions[0].id | https://openalex.org/I1315288073 |
| authorships[7].institutions[0].ror | https://ror.org/03s4khd80 |
| authorships[7].institutions[0].type | healthcare |
| authorships[7].institutions[0].lineage | https://openalex.org/I1315288073 |
| authorships[7].institutions[0].country_code | BE |
| authorships[7].institutions[0].display_name | Cliniques Universitaires Saint-Luc |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Nicole Revencu |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Cliniques universitaires Saint-Luc, University of Louvain |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.21203/rs.3.rs-4557690/v1 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Four Loss of Function Pathogenic Variants in ARHGAP29 in Non-Syndromic Cleft Lip and Palate |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T11374 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 0.9997000098228455 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1311 |
| primary_topic.subfield.display_name | Genetics |
| primary_topic.display_name | Cleft Lip and Palate Research |
| related_works | https://openalex.org/W2952784388, https://openalex.org/W2177105419, https://openalex.org/W3112847293, https://openalex.org/W2501856692, https://openalex.org/W2100847686, https://openalex.org/W2092849118, https://openalex.org/W2001105869, https://openalex.org/W2069150591, https://openalex.org/W4232163425, https://openalex.org/W4281651212 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.21203/rs.3.rs-4557690/v1 |
| best_oa_location.is_oa | True |
| best_oa_location.source | |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.21203/rs.3.rs-4557690/v1 |
| primary_location.id | doi:10.21203/rs.3.rs-4557690/v1 |
| primary_location.is_oa | True |
| primary_location.source | |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.21203/rs.3.rs-4557690/v1 |
| publication_date | 2024-07-15 |
| publication_year | 2024 |
| referenced_works | https://openalex.org/W1532587229, https://openalex.org/W2131036795, https://openalex.org/W2792928246, https://openalex.org/W2755420712, https://openalex.org/W2590327141, https://openalex.org/W2793871792, https://openalex.org/W2157928158, https://openalex.org/W4380853544, https://openalex.org/W1986819584, https://openalex.org/W2076687857, https://openalex.org/W2748230129, https://openalex.org/W3094012945, https://openalex.org/W2073846983, https://openalex.org/W2018130671, https://openalex.org/W2009181345, https://openalex.org/W1954195997, https://openalex.org/W2566174779, https://openalex.org/W2465904549, https://openalex.org/W3044440904, https://openalex.org/W2051978340, https://openalex.org/W2537562414, https://openalex.org/W3199859456, https://openalex.org/W3129351841, https://openalex.org/W1989851713, https://openalex.org/W2037761710, https://openalex.org/W2012359139, https://openalex.org/W2071805611, https://openalex.org/W1970413157, https://openalex.org/W2132340147, https://openalex.org/W2981777383, https://openalex.org/W1980958926, https://openalex.org/W2019096247, https://openalex.org/W3201649473, https://openalex.org/W1144139705, https://openalex.org/W4206799943, https://openalex.org/W2163344736, https://openalex.org/W2092550529, https://openalex.org/W4248277281, https://openalex.org/W2001593340, https://openalex.org/W3127861725, https://openalex.org/W2327251691 |
| referenced_works_count | 41 |
| abstract_inverted_index.7 | 46 |
| abstract_inverted_index.a | 52, 90, 110, 119 |
| abstract_inverted_index.29 | 39 |
| abstract_inverted_index.CL | 194 |
| abstract_inverted_index.ES | 70 |
| abstract_inverted_index.We | 50, 68 |
| abstract_inverted_index.an | 217 |
| abstract_inverted_index.as | 237 |
| abstract_inverted_index.be | 226, 240 |
| abstract_inverted_index.by | 65, 138, 142 |
| abstract_inverted_index.in | 45, 82, 84, 128, 167, 228, 242 |
| abstract_inverted_index.is | 11, 181, 187, 216 |
| abstract_inverted_index.it | 215, 238 |
| abstract_inverted_index.no | 59 |
| abstract_inverted_index.of | 4, 54, 97, 130, 148, 150, 175, 179, 210, 244 |
| abstract_inverted_index.or | 137, 141, 144 |
| abstract_inverted_index.to | 21, 94, 109, 126, 195 |
| abstract_inverted_index.224 | 55 |
| abstract_inverted_index.All | 122 |
| abstract_inverted_index.One | 88 |
| abstract_inverted_index.The | 1, 152, 162 |
| abstract_inverted_index.and | 71, 75, 114, 172, 212 |
| abstract_inverted_index.for | 57, 232 |
| abstract_inverted_index.had | 62 |
| abstract_inverted_index.has | 19, 41 |
| abstract_inverted_index.lip | 7 |
| abstract_inverted_index.may | 239 |
| abstract_inverted_index.not | 165, 188 |
| abstract_inverted_index.one | 117, 200 |
| abstract_inverted_index.the | 34, 95, 98, 115, 158, 168, 173, 207 |
| abstract_inverted_index.two | 103 |
| abstract_inverted_index.was | 89, 118 |
| abstract_inverted_index.(ES) | 18 |
| abstract_inverted_index.1.0, | 182 |
| abstract_inverted_index.CLP, | 198 |
| abstract_inverted_index.been | 42, 63 |
| abstract_inverted_index.four | 77, 85 |
| abstract_inverted_index.from | 192 |
| abstract_inverted_index.gene | 219 |
| abstract_inverted_index.high | 247 |
| abstract_inverted_index.lack | 157 |
| abstract_inverted_index.last | 116 |
| abstract_inverted_index.loss | 129, 174 |
| abstract_inverted_index.mRNA | 135 |
| abstract_inverted_index.show | 213 |
| abstract_inverted_index.that | 184, 214 |
| abstract_inverted_index.used | 69 |
| abstract_inverted_index.were | 104, 124, 164 |
| abstract_inverted_index.with | 25, 28, 48, 101, 199, 246 |
| abstract_inverted_index.(pLI) | 178 |
| abstract_inverted_index.Among | 31 |
| abstract_inverted_index.These | 204 |
| abstract_inverted_index.basis | 3 |
| abstract_inverted_index.cleft | 6 |
| abstract_inverted_index.exome | 16 |
| abstract_inverted_index.first | 99 |
| abstract_inverted_index.genes | 24 |
| abstract_inverted_index.novel | 78 |
| abstract_inverted_index.often | 27 |
| abstract_inverted_index.score | 177 |
| abstract_inverted_index.still | 12 |
| abstract_inverted_index.these | 32 |
| abstract_inverted_index.which | 58 |
| abstract_inverted_index.would | 156 |
| abstract_inverted_index.⁓2% | 243 |
| abstract_inverted_index.(88%). | 249 |
| abstract_inverted_index.GTPase | 36 |
| abstract_inverted_index.NsCL/P | 222 |
| abstract_inverted_index.NsCLPs | 56 |
| abstract_inverted_index.RhoGAP | 160 |
| abstract_inverted_index.and/or | 8 |
| abstract_inverted_index.cases, | 236 |
| abstract_inverted_index.codon, | 113 |
| abstract_inverted_index.cohort | 53 |
| abstract_inverted_index.decay, | 136 |
| abstract_inverted_index.either | 132 |
| abstract_inverted_index.extend | 206 |
| abstract_inverted_index.genes, | 33 |
| abstract_inverted_index.likely | 79 |
| abstract_inverted_index.palate | 9 |
| abstract_inverted_index.ranged | 191 |
| abstract_inverted_index.result | 127 |
| abstract_inverted_index.should | 225 |
| abstract_inverted_index.NsCL/P, | 26, 233 |
| abstract_inverted_index.NsCL/P. | 49 |
| abstract_inverted_index.allowed | 20 |
| abstract_inverted_index.control | 169 |
| abstract_inverted_index.domain. | 161 |
| abstract_inverted_index.genetic | 60, 230 |
| abstract_inverted_index.largely | 13 |
| abstract_inverted_index.leading | 93, 108 |
| abstract_inverted_index.mutated | 241 |
| abstract_inverted_index.present | 166 |
| abstract_inverted_index.protein | 38 |
| abstract_inverted_index.reduced | 29 |
| abstract_inverted_index.results | 205 |
| abstract_inverted_index.several | 23 |
| abstract_inverted_index.testing | 231 |
| abstract_inverted_index.through | 133 |
| abstract_inverted_index.variant | 73, 92 |
| abstract_inverted_index.(NsCL/P) | 10 |
| abstract_inverted_index.However, | 15 |
| abstract_inverted_index.abnormal | 145 |
| abstract_inverted_index.complete | 196 |
| abstract_inverted_index.familial | 235 |
| abstract_inverted_index.families | 47 |
| abstract_inverted_index.included | 227 |
| abstract_inverted_index.missense | 91 |
| abstract_inverted_index.mutation | 61, 202 |
| abstract_inverted_index.nonsense | 120 |
| abstract_inverted_index.patients | 245 |
| abstract_inverted_index.proteins | 155 |
| abstract_inverted_index.spectrum | 209 |
| abstract_inverted_index.testing. | 67 |
| abstract_inverted_index.unclear. | 14 |
| abstract_inverted_index.variable | 221 |
| abstract_inverted_index.variant. | 121 |
| abstract_inverted_index.variants | 81, 107, 123, 163 |
| abstract_inverted_index.associate | 22 |
| abstract_inverted_index.bilateral | 197 |
| abstract_inverted_index.carriers. | 203 |
| abstract_inverted_index.families. | 87 |
| abstract_inverted_index.filtering | 74 |
| abstract_inverted_index.function, | 131 |
| abstract_inverted_index.important | 159, 218 |
| abstract_inverted_index.microform | 193 |
| abstract_inverted_index.multiplex | 86 |
| abstract_inverted_index.predicted | 125 |
| abstract_inverted_index.premature | 111 |
| abstract_inverted_index.truncated | 139, 153 |
| abstract_inverted_index.N-terminal | 146 |
| abstract_inverted_index.Phenotypes | 190 |
| abstract_inverted_index.activating | 37 |
| abstract_inverted_index.databases, | 171 |
| abstract_inverted_index.diagnostic | 66, 229 |
| abstract_inverted_index.especially | 234 |
| abstract_inverted_index.frameshift | 106 |
| abstract_inverted_index.identified | 64, 76 |
| abstract_inverted_index.implicated | 44 |
| abstract_inverted_index.initiation | 147 |
| abstract_inverted_index.methionine | 100 |
| abstract_inverted_index.mutational | 208 |
| abstract_inverted_index.penetrance | 248 |
| abstract_inverted_index.population | 170 |
| abstract_inverted_index.previously | 43 |
| abstract_inverted_index.sequencing | 17 |
| abstract_inverted_index.suggesting | 183 |
| abstract_inverted_index.threonine, | 102 |
| abstract_inverted_index.tolerated. | 189 |
| abstract_inverted_index.unaffected | 201 |
| abstract_inverted_index.underlying | 220 |
| abstract_inverted_index.<italic>Rho | 35 |
| abstract_inverted_index.intolerance | 176 |
| abstract_inverted_index.penetrance. | 30 |
| abstract_inverted_index.phenotypes. | 223 |
| abstract_inverted_index.termination | 112 |
| abstract_inverted_index.translation | 149 |
| abstract_inverted_index.NMD-mediated | 134 |
| abstract_inverted_index.heterozygous | 105 |
| abstract_inverted_index.investigated | 51 |
| abstract_inverted_index.substitution | 96 |
| abstract_inverted_index.bioinformatic | 72 |
| abstract_inverted_index.non-syndromic | 5 |
| abstract_inverted_index.non-translation | 143 |
| abstract_inverted_index.haploinsufficiency | 186 |
| abstract_inverted_index.pathophysiological | 2 |
| abstract_inverted_index.(ARHGAP29)</italic> | 40 |
| abstract_inverted_index.pathogenic/pathologic | 80 |
| abstract_inverted_index.<title>Abstract</title> | 0 |
| abstract_inverted_index.<italic>ARHGAP29</italic> | 83, 154, 180, 185, 211, 224 |
| abstract_inverted_index.<italic>ARHGAP29</italic>, | 140 |
| abstract_inverted_index.<italic>ARHGAP29</italic>. | 151 |
| cited_by_percentile_year | |
| countries_distinct_count | 2 |
| institutions_distinct_count | 8 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/3 |
| sustainable_development_goals[0].score | 0.5600000023841858 |
| sustainable_development_goals[0].display_name | Good health and well-being |
| citation_normalized_percentile.value | 0.15294401 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |