From closed to open: three dynamic states of membrane-bound cytochrome P450 3A4 Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1007/s10822-025-00589-1
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1007/s10822-025-00589-1
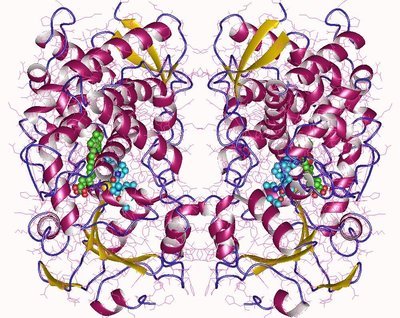
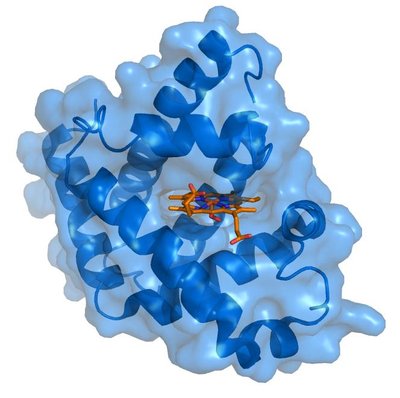
Cytochrome P450 3A4 (CYP3A4) is a membrane bound monooxygenase. It metabolizes the largest proportion of all orally ingested drugs. Ligands can enter and exit the enzyme through flexible tunnels, which co-determine CYP3A4’s ligand promiscuity. The flexibility can be represented by distinct conformational states of the enzyme. However, previous state definitions relied solely on crystal structures. We employed conventional molecular dynamics (cMD) simulations to sample the conformational space of CYP3A4. Five conformationally different crystal structures embedded in a membrane were simulated for 1 µs each. A Markov state model (MSM) coupled with spectral clustering (Robust Perron Cluster Analysis PCCA +) resulted in three distinct states: Two open conformations and an intermediate conformation. The tunnels inside CYP3A4 were calculated with CAVER3.0. Notably, we observed variations in bottleneck radii compared to those derived from crystallographic data. We want to point out the importance of simulations to characterize the dynamic behaviour. Moreover, we identified a mechanism, in which the membrane supports the opening of a tunnel. Therefore, CYP3A4 must be investigated in its membrane-bound state.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1007/s10822-025-00589-1
- https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdf
- OA Status
- hybrid
- Cited By
- 1
- References
- 78
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4408535875
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4408535875Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1007/s10822-025-00589-1Digital Object Identifier
- Title
-
From closed to open: three dynamic states of membrane-bound cytochrome P450 3A4Work title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-03-17Full publication date if available
- Authors
-
Vera A. Spanke, Valentin J. Egger-Hoerschinger, Veronika Ruzsányi, Klaus R. LiedlList of authors in order
- Landing page
-
https://doi.org/10.1007/s10822-025-00589-1Publisher landing page
- PDF URL
-
https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
hybridOpen access status per OpenAlex
- OA URL
-
https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdfDirect OA link when available
- Concepts
-
Molecular dynamics, Chemistry, Membrane, CYP3A4, Ligand (biochemistry), Crystallography, Stereochemistry, Chemical physics, Cytochrome P450, Computational chemistry, Enzyme, Biochemistry, ReceptorTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
1Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 1Per-year citation counts (last 5 years)
- References (count)
-
78Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4408535875 |
|---|---|
| doi | https://doi.org/10.1007/s10822-025-00589-1 |
| ids.doi | https://doi.org/10.1007/s10822-025-00589-1 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/40095179 |
| ids.openalex | https://openalex.org/W4408535875 |
| fwci | 4.98166126 |
| mesh[0].qualifier_ui | Q000201 |
| mesh[0].descriptor_ui | D002462 |
| mesh[0].is_major_topic | True |
| mesh[0].qualifier_name | enzymology |
| mesh[0].descriptor_name | Cell Membrane |
| mesh[1].qualifier_ui | Q000648 |
| mesh[1].descriptor_ui | D002462 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | ultrastructure |
| mesh[1].descriptor_name | Cell Membrane |
| mesh[2].qualifier_ui | Q000737 |
| mesh[2].descriptor_ui | D051544 |
| mesh[2].is_major_topic | True |
| mesh[2].qualifier_name | chemistry |
| mesh[2].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[3].qualifier_ui | Q000378 |
| mesh[3].descriptor_ui | D051544 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | metabolism |
| mesh[3].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[4].qualifier_ui | Q000648 |
| mesh[4].descriptor_ui | D051544 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | ultrastructure |
| mesh[4].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D057927 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Hydrophobic and Hydrophilic Interactions |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D008024 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Ligands |
| mesh[7].qualifier_ui | |
| mesh[7].descriptor_ui | D008390 |
| mesh[7].is_major_topic | False |
| mesh[7].qualifier_name | |
| mesh[7].descriptor_name | Markov Chains |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D056004 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Molecular Dynamics Simulation |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D011485 |
| mesh[9].is_major_topic | False |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Protein Binding |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D000072756 |
| mesh[10].is_major_topic | False |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Protein Conformation, alpha-Helical |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D018360 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Crystallography, X-Ray |
| mesh[12].qualifier_ui | Q000201 |
| mesh[12].descriptor_ui | D002462 |
| mesh[12].is_major_topic | True |
| mesh[12].qualifier_name | enzymology |
| mesh[12].descriptor_name | Cell Membrane |
| mesh[13].qualifier_ui | Q000648 |
| mesh[13].descriptor_ui | D002462 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | ultrastructure |
| mesh[13].descriptor_name | Cell Membrane |
| mesh[14].qualifier_ui | Q000737 |
| mesh[14].descriptor_ui | D051544 |
| mesh[14].is_major_topic | True |
| mesh[14].qualifier_name | chemistry |
| mesh[14].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[15].qualifier_ui | Q000378 |
| mesh[15].descriptor_ui | D051544 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | metabolism |
| mesh[15].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[16].qualifier_ui | Q000648 |
| mesh[16].descriptor_ui | D051544 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | ultrastructure |
| mesh[16].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D057927 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Hydrophobic and Hydrophilic Interactions |
| mesh[18].qualifier_ui | |
| mesh[18].descriptor_ui | D008024 |
| mesh[18].is_major_topic | False |
| mesh[18].qualifier_name | |
| mesh[18].descriptor_name | Ligands |
| mesh[19].qualifier_ui | |
| mesh[19].descriptor_ui | D008390 |
| mesh[19].is_major_topic | False |
| mesh[19].qualifier_name | |
| mesh[19].descriptor_name | Markov Chains |
| mesh[20].qualifier_ui | |
| mesh[20].descriptor_ui | D056004 |
| mesh[20].is_major_topic | False |
| mesh[20].qualifier_name | |
| mesh[20].descriptor_name | Molecular Dynamics Simulation |
| mesh[21].qualifier_ui | |
| mesh[21].descriptor_ui | D011485 |
| mesh[21].is_major_topic | False |
| mesh[21].qualifier_name | |
| mesh[21].descriptor_name | Protein Binding |
| mesh[22].qualifier_ui | |
| mesh[22].descriptor_ui | D000072756 |
| mesh[22].is_major_topic | False |
| mesh[22].qualifier_name | |
| mesh[22].descriptor_name | Protein Conformation, alpha-Helical |
| mesh[23].qualifier_ui | |
| mesh[23].descriptor_ui | D018360 |
| mesh[23].is_major_topic | False |
| mesh[23].qualifier_name | |
| mesh[23].descriptor_name | Crystallography, X-Ray |
| mesh[24].qualifier_ui | Q000737 |
| mesh[24].descriptor_ui | D051544 |
| mesh[24].is_major_topic | True |
| mesh[24].qualifier_name | chemistry |
| mesh[24].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[25].qualifier_ui | Q000378 |
| mesh[25].descriptor_ui | D051544 |
| mesh[25].is_major_topic | True |
| mesh[25].qualifier_name | metabolism |
| mesh[25].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[26].qualifier_ui | |
| mesh[26].descriptor_ui | D056004 |
| mesh[26].is_major_topic | True |
| mesh[26].qualifier_name | |
| mesh[26].descriptor_name | Molecular Dynamics Simulation |
| mesh[27].qualifier_ui | |
| mesh[27].descriptor_ui | D011487 |
| mesh[27].is_major_topic | True |
| mesh[27].qualifier_name | |
| mesh[27].descriptor_name | Protein Conformation |
| mesh[28].qualifier_ui | |
| mesh[28].descriptor_ui | D006801 |
| mesh[28].is_major_topic | False |
| mesh[28].qualifier_name | |
| mesh[28].descriptor_name | Humans |
| mesh[29].qualifier_ui | |
| mesh[29].descriptor_ui | D008024 |
| mesh[29].is_major_topic | False |
| mesh[29].qualifier_name | |
| mesh[29].descriptor_name | Ligands |
| mesh[30].qualifier_ui | |
| mesh[30].descriptor_ui | D018360 |
| mesh[30].is_major_topic | False |
| mesh[30].qualifier_name | |
| mesh[30].descriptor_name | Crystallography, X-Ray |
| mesh[31].qualifier_ui | |
| mesh[31].descriptor_ui | D008390 |
| mesh[31].is_major_topic | False |
| mesh[31].qualifier_name | |
| mesh[31].descriptor_name | Markov Chains |
| mesh[32].qualifier_ui | Q000378 |
| mesh[32].descriptor_ui | D002462 |
| mesh[32].is_major_topic | False |
| mesh[32].qualifier_name | metabolism |
| mesh[32].descriptor_name | Cell Membrane |
| mesh[33].qualifier_ui | Q000201 |
| mesh[33].descriptor_ui | D002462 |
| mesh[33].is_major_topic | False |
| mesh[33].qualifier_name | enzymology |
| mesh[33].descriptor_name | Cell Membrane |
| mesh[34].qualifier_ui | |
| mesh[34].descriptor_ui | D011485 |
| mesh[34].is_major_topic | False |
| mesh[34].qualifier_name | |
| mesh[34].descriptor_name | Protein Binding |
| mesh[35].qualifier_ui | Q000737 |
| mesh[35].descriptor_ui | D051544 |
| mesh[35].is_major_topic | True |
| mesh[35].qualifier_name | chemistry |
| mesh[35].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[36].qualifier_ui | Q000378 |
| mesh[36].descriptor_ui | D051544 |
| mesh[36].is_major_topic | True |
| mesh[36].qualifier_name | metabolism |
| mesh[36].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[37].qualifier_ui | |
| mesh[37].descriptor_ui | D056004 |
| mesh[37].is_major_topic | True |
| mesh[37].qualifier_name | |
| mesh[37].descriptor_name | Molecular Dynamics Simulation |
| mesh[38].qualifier_ui | |
| mesh[38].descriptor_ui | D011487 |
| mesh[38].is_major_topic | True |
| mesh[38].qualifier_name | |
| mesh[38].descriptor_name | Protein Conformation |
| mesh[39].qualifier_ui | |
| mesh[39].descriptor_ui | D006801 |
| mesh[39].is_major_topic | False |
| mesh[39].qualifier_name | |
| mesh[39].descriptor_name | Humans |
| mesh[40].qualifier_ui | |
| mesh[40].descriptor_ui | D008024 |
| mesh[40].is_major_topic | False |
| mesh[40].qualifier_name | |
| mesh[40].descriptor_name | Ligands |
| mesh[41].qualifier_ui | |
| mesh[41].descriptor_ui | D018360 |
| mesh[41].is_major_topic | False |
| mesh[41].qualifier_name | |
| mesh[41].descriptor_name | Crystallography, X-Ray |
| mesh[42].qualifier_ui | |
| mesh[42].descriptor_ui | D008390 |
| mesh[42].is_major_topic | False |
| mesh[42].qualifier_name | |
| mesh[42].descriptor_name | Markov Chains |
| mesh[43].qualifier_ui | Q000378 |
| mesh[43].descriptor_ui | D002462 |
| mesh[43].is_major_topic | False |
| mesh[43].qualifier_name | metabolism |
| mesh[43].descriptor_name | Cell Membrane |
| mesh[44].qualifier_ui | Q000201 |
| mesh[44].descriptor_ui | D002462 |
| mesh[44].is_major_topic | False |
| mesh[44].qualifier_name | enzymology |
| mesh[44].descriptor_name | Cell Membrane |
| mesh[45].qualifier_ui | |
| mesh[45].descriptor_ui | D011485 |
| mesh[45].is_major_topic | False |
| mesh[45].qualifier_name | |
| mesh[45].descriptor_name | Protein Binding |
| mesh[46].qualifier_ui | Q000737 |
| mesh[46].descriptor_ui | D051544 |
| mesh[46].is_major_topic | True |
| mesh[46].qualifier_name | chemistry |
| mesh[46].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[47].qualifier_ui | Q000378 |
| mesh[47].descriptor_ui | D051544 |
| mesh[47].is_major_topic | True |
| mesh[47].qualifier_name | metabolism |
| mesh[47].descriptor_name | Cytochrome P-450 CYP3A |
| mesh[48].qualifier_ui | |
| mesh[48].descriptor_ui | D056004 |
| mesh[48].is_major_topic | True |
| mesh[48].qualifier_name | |
| mesh[48].descriptor_name | Molecular Dynamics Simulation |
| mesh[49].qualifier_ui | |
| mesh[49].descriptor_ui | D011487 |
| mesh[49].is_major_topic | True |
| mesh[49].qualifier_name | |
| mesh[49].descriptor_name | Protein Conformation |
| type | article |
| title | From closed to open: three dynamic states of membrane-bound cytochrome P450 3A4 |
| awards[0].id | https://openalex.org/G7819292833 |
| awards[0].funder_id | https://openalex.org/F4320321181 |
| awards[0].display_name | Towards Personalised Medicine: use of volatile metabolites |
| awards[0].funder_award_id | P35312 |
| awards[0].funder_display_name | Austrian Science Fund |
| biblio.issue | 1 |
| biblio.volume | 39 |
| biblio.last_page | 12 |
| biblio.first_page | 12 |
| topics[0].id | https://openalex.org/T10375 |
| topics[0].field.id | https://openalex.org/fields/30 |
| topics[0].field.display_name | Pharmacology, Toxicology and Pharmaceutics |
| topics[0].score | 0.9998000264167786 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/3004 |
| topics[0].subfield.display_name | Pharmacology |
| topics[0].display_name | Pharmacogenetics and Drug Metabolism |
| topics[1].id | https://openalex.org/T10211 |
| topics[1].field.id | https://openalex.org/fields/17 |
| topics[1].field.display_name | Computer Science |
| topics[1].score | 0.9995999932289124 |
| topics[1].domain.id | https://openalex.org/domains/3 |
| topics[1].domain.display_name | Physical Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1703 |
| topics[1].subfield.display_name | Computational Theory and Mathematics |
| topics[1].display_name | Computational Drug Discovery Methods |
| topics[2].id | https://openalex.org/T10908 |
| topics[2].field.id | https://openalex.org/fields/16 |
| topics[2].field.display_name | Chemistry |
| topics[2].score | 0.9979000091552734 |
| topics[2].domain.id | https://openalex.org/domains/3 |
| topics[2].domain.display_name | Physical Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1607 |
| topics[2].subfield.display_name | Spectroscopy |
| topics[2].display_name | Analytical Chemistry and Chromatography |
| funders[0].id | https://openalex.org/F4320321181 |
| funders[0].ror | https://ror.org/013tf3c58 |
| funders[0].display_name | Austrian Science Fund |
| is_xpac | False |
| apc_list.value | 2890 |
| apc_list.currency | EUR |
| apc_list.value_usd | 3690 |
| apc_paid.value | 2890 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 3690 |
| concepts[0].id | https://openalex.org/C59593255 |
| concepts[0].level | 2 |
| concepts[0].score | 0.677417516708374 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q901663 |
| concepts[0].display_name | Molecular dynamics |
| concepts[1].id | https://openalex.org/C185592680 |
| concepts[1].level | 0 |
| concepts[1].score | 0.5729209184646606 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q2329 |
| concepts[1].display_name | Chemistry |
| concepts[2].id | https://openalex.org/C41625074 |
| concepts[2].level | 2 |
| concepts[2].score | 0.45803430676460266 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q176088 |
| concepts[2].display_name | Membrane |
| concepts[3].id | https://openalex.org/C109650736 |
| concepts[3].level | 4 |
| concepts[3].score | 0.4434959292411804 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q424440 |
| concepts[3].display_name | CYP3A4 |
| concepts[4].id | https://openalex.org/C116569031 |
| concepts[4].level | 3 |
| concepts[4].score | 0.42455482482910156 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q899107 |
| concepts[4].display_name | Ligand (biochemistry) |
| concepts[5].id | https://openalex.org/C8010536 |
| concepts[5].level | 1 |
| concepts[5].score | 0.3666074573993683 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q160398 |
| concepts[5].display_name | Crystallography |
| concepts[6].id | https://openalex.org/C71240020 |
| concepts[6].level | 1 |
| concepts[6].score | 0.34485098719596863 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q186011 |
| concepts[6].display_name | Stereochemistry |
| concepts[7].id | https://openalex.org/C159467904 |
| concepts[7].level | 1 |
| concepts[7].score | 0.3263806700706482 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q2001702 |
| concepts[7].display_name | Chemical physics |
| concepts[8].id | https://openalex.org/C526171541 |
| concepts[8].level | 3 |
| concepts[8].score | 0.2876339554786682 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q407693 |
| concepts[8].display_name | Cytochrome P450 |
| concepts[9].id | https://openalex.org/C147597530 |
| concepts[9].level | 1 |
| concepts[9].score | 0.2490626871585846 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q369472 |
| concepts[9].display_name | Computational chemistry |
| concepts[10].id | https://openalex.org/C181199279 |
| concepts[10].level | 2 |
| concepts[10].score | 0.23720061779022217 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q8047 |
| concepts[10].display_name | Enzyme |
| concepts[11].id | https://openalex.org/C55493867 |
| concepts[11].level | 1 |
| concepts[11].score | 0.07736346125602722 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7094 |
| concepts[11].display_name | Biochemistry |
| concepts[12].id | https://openalex.org/C170493617 |
| concepts[12].level | 2 |
| concepts[12].score | 0.0 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q208467 |
| concepts[12].display_name | Receptor |
| keywords[0].id | https://openalex.org/keywords/molecular-dynamics |
| keywords[0].score | 0.677417516708374 |
| keywords[0].display_name | Molecular dynamics |
| keywords[1].id | https://openalex.org/keywords/chemistry |
| keywords[1].score | 0.5729209184646606 |
| keywords[1].display_name | Chemistry |
| keywords[2].id | https://openalex.org/keywords/membrane |
| keywords[2].score | 0.45803430676460266 |
| keywords[2].display_name | Membrane |
| keywords[3].id | https://openalex.org/keywords/cyp3a4 |
| keywords[3].score | 0.4434959292411804 |
| keywords[3].display_name | CYP3A4 |
| keywords[4].id | https://openalex.org/keywords/ligand |
| keywords[4].score | 0.42455482482910156 |
| keywords[4].display_name | Ligand (biochemistry) |
| keywords[5].id | https://openalex.org/keywords/crystallography |
| keywords[5].score | 0.3666074573993683 |
| keywords[5].display_name | Crystallography |
| keywords[6].id | https://openalex.org/keywords/stereochemistry |
| keywords[6].score | 0.34485098719596863 |
| keywords[6].display_name | Stereochemistry |
| keywords[7].id | https://openalex.org/keywords/chemical-physics |
| keywords[7].score | 0.3263806700706482 |
| keywords[7].display_name | Chemical physics |
| keywords[8].id | https://openalex.org/keywords/cytochrome-p450 |
| keywords[8].score | 0.2876339554786682 |
| keywords[8].display_name | Cytochrome P450 |
| keywords[9].id | https://openalex.org/keywords/computational-chemistry |
| keywords[9].score | 0.2490626871585846 |
| keywords[9].display_name | Computational chemistry |
| keywords[10].id | https://openalex.org/keywords/enzyme |
| keywords[10].score | 0.23720061779022217 |
| keywords[10].display_name | Enzyme |
| keywords[11].id | https://openalex.org/keywords/biochemistry |
| keywords[11].score | 0.07736346125602722 |
| keywords[11].display_name | Biochemistry |
| language | en |
| locations[0].id | doi:10.1007/s10822-025-00589-1 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S64621741 |
| locations[0].source.issn | 0920-654X, 1573-4951 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | 0920-654X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Journal of Computer-Aided Molecular Design |
| locations[0].source.host_organization | https://openalex.org/P4310319900 |
| locations[0].source.host_organization_name | Springer Science+Business Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| locations[0].source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Journal of Computer-Aided Molecular Design |
| locations[0].landing_page_url | https://doi.org/10.1007/s10822-025-00589-1 |
| locations[1].id | pmid:40095179 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Journal of computer-aided molecular design |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/40095179 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:11913904 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | other-oa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/other-oa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | J Comput Aided Mol Des |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/11913904 |
| indexed_in | crossref, pubmed |
| authorships[0].author.id | https://openalex.org/A5115939286 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Vera A. Spanke |
| authorships[0].countries | AT |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I190249584 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Theoretical Chemistry, Universität Innsbruck, Innsbruck, Austria |
| authorships[0].institutions[0].id | https://openalex.org/I190249584 |
| authorships[0].institutions[0].ror | https://ror.org/054pv6659 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I190249584 |
| authorships[0].institutions[0].country_code | AT |
| authorships[0].institutions[0].display_name | Universität Innsbruck |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Vera A. Spanke |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Theoretical Chemistry, Universität Innsbruck, Innsbruck, Austria |
| authorships[1].author.id | https://openalex.org/A5116664685 |
| authorships[1].author.orcid | |
| authorships[1].author.display_name | Valentin J. Egger-Hoerschinger |
| authorships[1].countries | AT |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I190249584 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Theoretical Chemistry, Universität Innsbruck, Innsbruck, Austria |
| authorships[1].institutions[0].id | https://openalex.org/I190249584 |
| authorships[1].institutions[0].ror | https://ror.org/054pv6659 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I190249584 |
| authorships[1].institutions[0].country_code | AT |
| authorships[1].institutions[0].display_name | Universität Innsbruck |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Valentin J. Egger-Hoerschinger |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Theoretical Chemistry, Universität Innsbruck, Innsbruck, Austria |
| authorships[2].author.id | https://openalex.org/A5069465132 |
| authorships[2].author.orcid | https://orcid.org/0000-0001-9631-364X |
| authorships[2].author.display_name | Veronika Ruzsányi |
| authorships[2].countries | AT |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I190249584 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Breath Research, Universität Innsbruck, Innsbruck, Austria |
| authorships[2].institutions[0].id | https://openalex.org/I190249584 |
| authorships[2].institutions[0].ror | https://ror.org/054pv6659 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I190249584 |
| authorships[2].institutions[0].country_code | AT |
| authorships[2].institutions[0].display_name | Universität Innsbruck |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Veronika Ruzsanyi |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Breath Research, Universität Innsbruck, Innsbruck, Austria |
| authorships[3].author.id | https://openalex.org/A5028405436 |
| authorships[3].author.orcid | https://orcid.org/0000-0002-0985-2299 |
| authorships[3].author.display_name | Klaus R. Liedl |
| authorships[3].countries | AT |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I190249584 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Theoretical Chemistry, Universität Innsbruck, Innsbruck, Austria |
| authorships[3].institutions[0].id | https://openalex.org/I190249584 |
| authorships[3].institutions[0].ror | https://ror.org/054pv6659 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I190249584 |
| authorships[3].institutions[0].country_code | AT |
| authorships[3].institutions[0].display_name | Universität Innsbruck |
| authorships[3].author_position | last |
| authorships[3].raw_author_name | Klaus R. Liedl |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Theoretical Chemistry, Universität Innsbruck, Innsbruck, Austria |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdf |
| open_access.oa_status | hybrid |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | From closed to open: three dynamic states of membrane-bound cytochrome P450 3A4 |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-07T23:20:04.922697 |
| primary_topic.id | https://openalex.org/T10375 |
| primary_topic.field.id | https://openalex.org/fields/30 |
| primary_topic.field.display_name | Pharmacology, Toxicology and Pharmaceutics |
| primary_topic.score | 0.9998000264167786 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/3004 |
| primary_topic.subfield.display_name | Pharmacology |
| primary_topic.display_name | Pharmacogenetics and Drug Metabolism |
| related_works | https://openalex.org/W2150391754, https://openalex.org/W2064710306, https://openalex.org/W1997484252, https://openalex.org/W1979156653, https://openalex.org/W2249456921, https://openalex.org/W2184834470, https://openalex.org/W2334502792, https://openalex.org/W2316398224, https://openalex.org/W4281654480, https://openalex.org/W2001372703 |
| cited_by_count | 1 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 1 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1007/s10822-025-00589-1 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S64621741 |
| best_oa_location.source.issn | 0920-654X, 1573-4951 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | 0920-654X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Journal of Computer-Aided Molecular Design |
| best_oa_location.source.host_organization | https://openalex.org/P4310319900 |
| best_oa_location.source.host_organization_name | Springer Science+Business Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| best_oa_location.source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Journal of Computer-Aided Molecular Design |
| best_oa_location.landing_page_url | https://doi.org/10.1007/s10822-025-00589-1 |
| primary_location.id | doi:10.1007/s10822-025-00589-1 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S64621741 |
| primary_location.source.issn | 0920-654X, 1573-4951 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | 0920-654X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Journal of Computer-Aided Molecular Design |
| primary_location.source.host_organization | https://openalex.org/P4310319900 |
| primary_location.source.host_organization_name | Springer Science+Business Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310319900, https://openalex.org/P4310319965 |
| primary_location.source.host_organization_lineage_names | Springer Science+Business Media, Springer Nature |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://link.springer.com/content/pdf/10.1007/s10822-025-00589-1.pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Journal of Computer-Aided Molecular Design |
| primary_location.landing_page_url | https://doi.org/10.1007/s10822-025-00589-1 |
| publication_date | 2025-03-17 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W4307266448, https://openalex.org/W2127059331, https://openalex.org/W2005328285, https://openalex.org/W2113524447, https://openalex.org/W2907653052, https://openalex.org/W2798194035, https://openalex.org/W1987275848, https://openalex.org/W2081280346, https://openalex.org/W2069518737, https://openalex.org/W2146630606, https://openalex.org/W2953039797, https://openalex.org/W2149573717, https://openalex.org/W4385383151, https://openalex.org/W2296756275, https://openalex.org/W4362637453, https://openalex.org/W3190153844, https://openalex.org/W1999848286, https://openalex.org/W4402607711, https://openalex.org/W3134454869, https://openalex.org/W2161069015, https://openalex.org/W2725934788, https://openalex.org/W2130479394, https://openalex.org/W2327129143, https://openalex.org/W2270260897, https://openalex.org/W1963968986, https://openalex.org/W2785939655, https://openalex.org/W2137825366, https://openalex.org/W4395682877, https://openalex.org/W2587649658, https://openalex.org/W2401018553, https://openalex.org/W1999698423, https://openalex.org/W2749132107, https://openalex.org/W2226825552, https://openalex.org/W2014919794, https://openalex.org/W2130983257, https://openalex.org/W2147472643, https://openalex.org/W2090569544, https://openalex.org/W2773174471, https://openalex.org/W3177828909, https://openalex.org/W4283326616, https://openalex.org/W2311078281, https://openalex.org/W2160907977, https://openalex.org/W1990771438, https://openalex.org/W2140831051, https://openalex.org/W1976499671, https://openalex.org/W2404280981, https://openalex.org/W2323104785, https://openalex.org/W2064867387, https://openalex.org/W2134762386, https://openalex.org/W2313116527, https://openalex.org/W2017001642, https://openalex.org/W2035266068, https://openalex.org/W2103945336, https://openalex.org/W2110479326, https://openalex.org/W2056211671, https://openalex.org/W2121939683, https://openalex.org/W2143592395, https://openalex.org/W2012029966, https://openalex.org/W1967005434, https://openalex.org/W2151370125, https://openalex.org/W2332712348, https://openalex.org/W3138914381, https://openalex.org/W2944202352, https://openalex.org/W2134967712, https://openalex.org/W2011301426, https://openalex.org/W4298850410, https://openalex.org/W2140673705, https://openalex.org/W2168269595, https://openalex.org/W2038530231, https://openalex.org/W2010177770, https://openalex.org/W2530590278, https://openalex.org/W2916087834, https://openalex.org/W423419411, https://openalex.org/W2050641174, https://openalex.org/W2519280988, https://openalex.org/W2329705955, https://openalex.org/W3100810942, https://openalex.org/W4404446318 |
| referenced_works_count | 78 |
| abstract_inverted_index.1 | 82 |
| abstract_inverted_index.A | 85 |
| abstract_inverted_index.a | 6, 77, 151, 161 |
| abstract_inverted_index.+) | 99 |
| abstract_inverted_index.It | 10 |
| abstract_inverted_index.We | 56, 134 |
| abstract_inverted_index.an | 109 |
| abstract_inverted_index.be | 38, 166 |
| abstract_inverted_index.by | 40 |
| abstract_inverted_index.in | 76, 101, 124, 153, 168 |
| abstract_inverted_index.is | 5 |
| abstract_inverted_index.of | 15, 44, 68, 141, 160 |
| abstract_inverted_index.on | 53 |
| abstract_inverted_index.to | 63, 128, 136, 143 |
| abstract_inverted_index.we | 121, 149 |
| abstract_inverted_index.3A4 | 3 |
| abstract_inverted_index.The | 35, 112 |
| abstract_inverted_index.Two | 105 |
| abstract_inverted_index.all | 16 |
| abstract_inverted_index.and | 23, 108 |
| abstract_inverted_index.can | 21, 37 |
| abstract_inverted_index.for | 81 |
| abstract_inverted_index.its | 169 |
| abstract_inverted_index.out | 138 |
| abstract_inverted_index.the | 12, 25, 45, 65, 139, 145, 155, 158 |
| abstract_inverted_index.µs | 83 |
| abstract_inverted_index.Five | 70 |
| abstract_inverted_index.P450 | 2 |
| abstract_inverted_index.PCCA | 98 |
| abstract_inverted_index.exit | 24 |
| abstract_inverted_index.from | 131 |
| abstract_inverted_index.must | 165 |
| abstract_inverted_index.open | 106 |
| abstract_inverted_index.want | 135 |
| abstract_inverted_index.were | 79, 116 |
| abstract_inverted_index.with | 91, 118 |
| abstract_inverted_index.(MSM) | 89 |
| abstract_inverted_index.(cMD) | 61 |
| abstract_inverted_index.bound | 8 |
| abstract_inverted_index.data. | 133 |
| abstract_inverted_index.each. | 84 |
| abstract_inverted_index.enter | 22 |
| abstract_inverted_index.model | 88 |
| abstract_inverted_index.point | 137 |
| abstract_inverted_index.radii | 126 |
| abstract_inverted_index.space | 67 |
| abstract_inverted_index.state | 49, 87 |
| abstract_inverted_index.those | 129 |
| abstract_inverted_index.three | 102 |
| abstract_inverted_index.which | 30, 154 |
| abstract_inverted_index.CYP3A4 | 115, 164 |
| abstract_inverted_index.Markov | 86 |
| abstract_inverted_index.Perron | 95 |
| abstract_inverted_index.drugs. | 19 |
| abstract_inverted_index.enzyme | 26 |
| abstract_inverted_index.inside | 114 |
| abstract_inverted_index.ligand | 33 |
| abstract_inverted_index.orally | 17 |
| abstract_inverted_index.relied | 51 |
| abstract_inverted_index.sample | 64 |
| abstract_inverted_index.solely | 52 |
| abstract_inverted_index.state. | 171 |
| abstract_inverted_index.states | 43 |
| abstract_inverted_index.(Robust | 94 |
| abstract_inverted_index.CYP3A4. | 69 |
| abstract_inverted_index.Cluster | 96 |
| abstract_inverted_index.Ligands | 20 |
| abstract_inverted_index.coupled | 90 |
| abstract_inverted_index.crystal | 54, 73 |
| abstract_inverted_index.derived | 130 |
| abstract_inverted_index.dynamic | 146 |
| abstract_inverted_index.enzyme. | 46 |
| abstract_inverted_index.largest | 13 |
| abstract_inverted_index.opening | 159 |
| abstract_inverted_index.states: | 104 |
| abstract_inverted_index.through | 27 |
| abstract_inverted_index.tunnel. | 162 |
| abstract_inverted_index.tunnels | 113 |
| abstract_inverted_index.(CYP3A4) | 4 |
| abstract_inverted_index.Abstract | 0 |
| abstract_inverted_index.Analysis | 97 |
| abstract_inverted_index.However, | 47 |
| abstract_inverted_index.Notably, | 120 |
| abstract_inverted_index.compared | 127 |
| abstract_inverted_index.distinct | 41, 103 |
| abstract_inverted_index.dynamics | 60 |
| abstract_inverted_index.embedded | 75 |
| abstract_inverted_index.employed | 57 |
| abstract_inverted_index.flexible | 28 |
| abstract_inverted_index.ingested | 18 |
| abstract_inverted_index.membrane | 7, 78, 156 |
| abstract_inverted_index.observed | 122 |
| abstract_inverted_index.previous | 48 |
| abstract_inverted_index.resulted | 100 |
| abstract_inverted_index.spectral | 92 |
| abstract_inverted_index.supports | 157 |
| abstract_inverted_index.tunnels, | 29 |
| abstract_inverted_index.CAVER3.0. | 119 |
| abstract_inverted_index.Moreover, | 148 |
| abstract_inverted_index.different | 72 |
| abstract_inverted_index.molecular | 59 |
| abstract_inverted_index.simulated | 80 |
| abstract_inverted_index.CYP3A4’s | 32 |
| abstract_inverted_index.Cytochrome | 1 |
| abstract_inverted_index.Therefore, | 163 |
| abstract_inverted_index.behaviour. | 147 |
| abstract_inverted_index.bottleneck | 125 |
| abstract_inverted_index.calculated | 117 |
| abstract_inverted_index.clustering | 93 |
| abstract_inverted_index.identified | 150 |
| abstract_inverted_index.importance | 140 |
| abstract_inverted_index.mechanism, | 152 |
| abstract_inverted_index.proportion | 14 |
| abstract_inverted_index.structures | 74 |
| abstract_inverted_index.variations | 123 |
| abstract_inverted_index.definitions | 50 |
| abstract_inverted_index.flexibility | 36 |
| abstract_inverted_index.metabolizes | 11 |
| abstract_inverted_index.represented | 39 |
| abstract_inverted_index.simulations | 62, 142 |
| abstract_inverted_index.structures. | 55 |
| abstract_inverted_index.characterize | 144 |
| abstract_inverted_index.co-determine | 31 |
| abstract_inverted_index.conventional | 58 |
| abstract_inverted_index.intermediate | 110 |
| abstract_inverted_index.investigated | 167 |
| abstract_inverted_index.promiscuity. | 34 |
| abstract_inverted_index.conformation. | 111 |
| abstract_inverted_index.conformations | 107 |
| abstract_inverted_index.conformational | 42, 66 |
| abstract_inverted_index.membrane-bound | 170 |
| abstract_inverted_index.monooxygenase. | 9 |
| abstract_inverted_index.conformationally | 71 |
| abstract_inverted_index.crystallographic | 132 |
| cited_by_percentile_year.max | 95 |
| cited_by_percentile_year.min | 91 |
| countries_distinct_count | 1 |
| institutions_distinct_count | 4 |
| citation_normalized_percentile.value | 0.8406189 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | True |