Functional Mapping of Phenotypic Plasticity of Staphylococcus aureus Under Vancomycin Pressure Article Swipe
 YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3389/fmicb.2021.696730
YOU?
·
· 2021
· Open Access
·
· DOI: https://doi.org/10.3389/fmicb.2021.696730
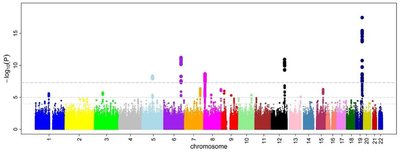
Phenotypic plasticity is the exhibition of various phenotypic traits produced by a single genotype in response to environmental changes, enabling organisms to adapt to environmental changes by maintaining growth and reproduction. Despite its significance in evolutionary studies, we still know little about the genetic control of phenotypic plasticity. In this study, we designed and conducted a genome-wide association study (GWAS) to reveal genetic architecture of how Staphylococcus aureus strains respond to increasing concentrations of vancomycin (0, 2, 4, and 6 μg/mL) in a time course. We implemented functional mapping, a dynamic model for genetic mapping using longitudinal data, to map specific loci that mediate the growth trajectories of abundance of vancomycin-exposed S. aureus strains. 78 significant single nucleotide polymorphisms were identified following analysis of the whole growth and development process, and seven genes might play a pivotal role in governing phenotypic plasticity to the pressure of vancomycin. These seven genes, SAOUHSC_00020 ( walR ), SAOUHSC_00176, SAOUHSC_00544 ( sdrC ), SAOUHSC_02998, SAOUHSC_00025, SAOUHSC_00169, and SAOUHSC_02023, were found to help S. aureus regulate antibiotic pressure. Our dynamic gene mapping technique provides a tool for dissecting the phenotypic plasticity mechanisms of S. aureus under vancomycin pressure, emphasizing the feasibility and potential of functional mapping in the study of bacterial phenotypic plasticity.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.3389/fmicb.2021.696730
- https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdf
- OA Status
- gold
- Cited By
- 5
- References
- 55
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W3197280922
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W3197280922Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.3389/fmicb.2021.696730Digital Object Identifier
- Title
-
Functional Mapping of Phenotypic Plasticity of Staphylococcus aureus Under Vancomycin PressureWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2021Year of publication
- Publication date
-
2021-09-09Full publication date if available
- Authors
-
Dengcheng Yang, Xuyang Zheng, Libo Jiang, Meixia Ye, Xiaoqing He, Yi Jin, Rongling WuList of authors in order
- Landing page
-
https://doi.org/10.3389/fmicb.2021.696730Publisher landing page
- PDF URL
-
https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdfDirect OA link when available
- Concepts
-
Biology, Phenotype, Phenotypic plasticity, Staphylococcus aureus, Genetic architecture, Genetics, Vancomycin, Genome-wide association study, Gene, Single-nucleotide polymorphism, Genotype, BacteriaTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
5Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 2, 2024: 2, 2022: 1Per-year citation counts (last 5 years)
- References (count)
-
55Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W3197280922 |
|---|---|
| doi | https://doi.org/10.3389/fmicb.2021.696730 |
| ids.doi | https://doi.org/10.3389/fmicb.2021.696730 |
| ids.mag | 3197280922 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/34566908 |
| ids.openalex | https://openalex.org/W3197280922 |
| fwci | 0.2194311 |
| type | article |
| title | Functional Mapping of Phenotypic Plasticity of Staphylococcus aureus Under Vancomycin Pressure |
| biblio.issue | |
| biblio.volume | 12 |
| biblio.last_page | 696730 |
| biblio.first_page | 696730 |
| topics[0].id | https://openalex.org/T10195 |
| topics[0].field.id | https://openalex.org/fields/27 |
| topics[0].field.display_name | Medicine |
| topics[0].score | 0.9970999956130981 |
| topics[0].domain.id | https://openalex.org/domains/4 |
| topics[0].domain.display_name | Health Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2725 |
| topics[0].subfield.display_name | Infectious Diseases |
| topics[0].display_name | Antimicrobial Resistance in Staphylococcus |
| topics[1].id | https://openalex.org/T10120 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9947999715805054 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1311 |
| topics[1].subfield.display_name | Genetics |
| topics[1].display_name | Bacterial Genetics and Biotechnology |
| topics[2].id | https://openalex.org/T10521 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9930999875068665 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | RNA and protein synthesis mechanisms |
| is_xpac | False |
| apc_list.value | 2950 |
| apc_list.currency | USD |
| apc_list.value_usd | 2950 |
| apc_paid.value | 2950 |
| apc_paid.currency | USD |
| apc_paid.value_usd | 2950 |
| concepts[0].id | https://openalex.org/C86803240 |
| concepts[0].level | 0 |
| concepts[0].score | 0.8085997104644775 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[0].display_name | Biology |
| concepts[1].id | https://openalex.org/C127716648 |
| concepts[1].level | 3 |
| concepts[1].score | 0.6550567746162415 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q104053 |
| concepts[1].display_name | Phenotype |
| concepts[2].id | https://openalex.org/C62277055 |
| concepts[2].level | 2 |
| concepts[2].score | 0.6334211230278015 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q295130 |
| concepts[2].display_name | Phenotypic plasticity |
| concepts[3].id | https://openalex.org/C2779489039 |
| concepts[3].level | 3 |
| concepts[3].score | 0.6051746010780334 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q188121 |
| concepts[3].display_name | Staphylococcus aureus |
| concepts[4].id | https://openalex.org/C9287583 |
| concepts[4].level | 4 |
| concepts[4].score | 0.46645206212997437 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q5532883 |
| concepts[4].display_name | Genetic architecture |
| concepts[5].id | https://openalex.org/C54355233 |
| concepts[5].level | 1 |
| concepts[5].score | 0.4594477415084839 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[5].display_name | Genetics |
| concepts[6].id | https://openalex.org/C2778980435 |
| concepts[6].level | 4 |
| concepts[6].score | 0.4563385546207428 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q424027 |
| concepts[6].display_name | Vancomycin |
| concepts[7].id | https://openalex.org/C106208931 |
| concepts[7].level | 5 |
| concepts[7].score | 0.4444258511066437 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[7].display_name | Genome-wide association study |
| concepts[8].id | https://openalex.org/C104317684 |
| concepts[8].level | 2 |
| concepts[8].score | 0.42697271704673767 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[8].display_name | Gene |
| concepts[9].id | https://openalex.org/C153209595 |
| concepts[9].level | 4 |
| concepts[9].score | 0.4104973375797272 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[9].display_name | Single-nucleotide polymorphism |
| concepts[10].id | https://openalex.org/C135763542 |
| concepts[10].level | 3 |
| concepts[10].score | 0.3555441200733185 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[10].display_name | Genotype |
| concepts[11].id | https://openalex.org/C523546767 |
| concepts[11].level | 2 |
| concepts[11].score | 0.15426185727119446 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q10876 |
| concepts[11].display_name | Bacteria |
| keywords[0].id | https://openalex.org/keywords/biology |
| keywords[0].score | 0.8085997104644775 |
| keywords[0].display_name | Biology |
| keywords[1].id | https://openalex.org/keywords/phenotype |
| keywords[1].score | 0.6550567746162415 |
| keywords[1].display_name | Phenotype |
| keywords[2].id | https://openalex.org/keywords/phenotypic-plasticity |
| keywords[2].score | 0.6334211230278015 |
| keywords[2].display_name | Phenotypic plasticity |
| keywords[3].id | https://openalex.org/keywords/staphylococcus-aureus |
| keywords[3].score | 0.6051746010780334 |
| keywords[3].display_name | Staphylococcus aureus |
| keywords[4].id | https://openalex.org/keywords/genetic-architecture |
| keywords[4].score | 0.46645206212997437 |
| keywords[4].display_name | Genetic architecture |
| keywords[5].id | https://openalex.org/keywords/genetics |
| keywords[5].score | 0.4594477415084839 |
| keywords[5].display_name | Genetics |
| keywords[6].id | https://openalex.org/keywords/vancomycin |
| keywords[6].score | 0.4563385546207428 |
| keywords[6].display_name | Vancomycin |
| keywords[7].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[7].score | 0.4444258511066437 |
| keywords[7].display_name | Genome-wide association study |
| keywords[8].id | https://openalex.org/keywords/gene |
| keywords[8].score | 0.42697271704673767 |
| keywords[8].display_name | Gene |
| keywords[9].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[9].score | 0.4104973375797272 |
| keywords[9].display_name | Single-nucleotide polymorphism |
| keywords[10].id | https://openalex.org/keywords/genotype |
| keywords[10].score | 0.3555441200733185 |
| keywords[10].display_name | Genotype |
| keywords[11].id | https://openalex.org/keywords/bacteria |
| keywords[11].score | 0.15426185727119446 |
| keywords[11].display_name | Bacteria |
| language | en |
| locations[0].id | doi:10.3389/fmicb.2021.696730 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2484704948 |
| locations[0].source.issn | 1664-302X |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 1664-302X |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Frontiers in Microbiology |
| locations[0].source.host_organization | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_name | Frontiers Media |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320527 |
| locations[0].source.host_organization_lineage_names | Frontiers Media |
| locations[0].license | cc-by |
| locations[0].pdf_url | https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdf |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Frontiers in Microbiology |
| locations[0].landing_page_url | https://doi.org/10.3389/fmicb.2021.696730 |
| locations[1].id | pmid:34566908 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Frontiers in microbiology |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/34566908 |
| locations[2].id | pmh:oai:doaj.org/article:02f90350849a490eb8dc405addd5eeb7 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S4306401280 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | DOAJ (DOAJ: Directory of Open Access Journals) |
| locations[2].source.host_organization | |
| locations[2].source.host_organization_name | |
| locations[2].license | cc-by-sa |
| locations[2].pdf_url | |
| locations[2].version | submittedVersion |
| locations[2].raw_type | article |
| locations[2].license_id | https://openalex.org/licenses/cc-by-sa |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Frontiers in Microbiology, Vol 12 (2021) |
| locations[2].landing_page_url | https://doaj.org/article/02f90350849a490eb8dc405addd5eeb7 |
| locations[3].id | pmh:oai:europepmc.org:7349174 |
| locations[3].is_oa | True |
| locations[3].source.id | https://openalex.org/S4306400806 |
| locations[3].source.issn | |
| locations[3].source.type | repository |
| locations[3].source.is_oa | False |
| locations[3].source.issn_l | |
| locations[3].source.is_core | False |
| locations[3].source.is_in_doaj | False |
| locations[3].source.display_name | Europe PMC (PubMed Central) |
| locations[3].source.host_organization | https://openalex.org/I1303153112 |
| locations[3].source.host_organization_name | European Bioinformatics Institute |
| locations[3].source.host_organization_lineage | https://openalex.org/I1303153112 |
| locations[3].license | other-oa |
| locations[3].pdf_url | |
| locations[3].version | submittedVersion |
| locations[3].raw_type | Text |
| locations[3].license_id | https://openalex.org/licenses/other-oa |
| locations[3].is_accepted | False |
| locations[3].is_published | False |
| locations[3].raw_source_name | |
| locations[3].landing_page_url | http://europepmc.org/pmc/articles/PMC8458881 |
| locations[4].id | pmh:oai:pubmedcentral.nih.gov:8458881 |
| locations[4].is_oa | True |
| locations[4].source.id | https://openalex.org/S2764455111 |
| locations[4].source.issn | |
| locations[4].source.type | repository |
| locations[4].source.is_oa | False |
| locations[4].source.issn_l | |
| locations[4].source.is_core | False |
| locations[4].source.is_in_doaj | False |
| locations[4].source.display_name | PubMed Central |
| locations[4].source.host_organization | https://openalex.org/I1299303238 |
| locations[4].source.host_organization_name | National Institutes of Health |
| locations[4].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[4].license | other-oa |
| locations[4].pdf_url | |
| locations[4].version | submittedVersion |
| locations[4].raw_type | Text |
| locations[4].license_id | https://openalex.org/licenses/other-oa |
| locations[4].is_accepted | False |
| locations[4].is_published | False |
| locations[4].raw_source_name | Front Microbiol |
| locations[4].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/8458881 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5036300046 |
| authorships[0].author.orcid | https://orcid.org/0000-0002-5879-1998 |
| authorships[0].author.display_name | Dengcheng Yang |
| authorships[0].countries | CN |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[0].affiliations[0].raw_affiliation_string | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[0].institutions[0].id | https://openalex.org/I31683504 |
| authorships[0].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[0].institutions[0].country_code | CN |
| authorships[0].institutions[0].display_name | Beijing Forestry University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Dengcheng Yang |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[1].author.id | https://openalex.org/A5101958014 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-1614-8029 |
| authorships[1].author.display_name | Xuyang Zheng |
| authorships[1].countries | CN |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[1].affiliations[0].raw_affiliation_string | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[1].institutions[0].id | https://openalex.org/I31683504 |
| authorships[1].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[1].institutions[0].type | education |
| authorships[1].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[1].institutions[0].country_code | CN |
| authorships[1].institutions[0].display_name | Beijing Forestry University |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Xuyang Zheng |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[2].author.id | https://openalex.org/A5020151036 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-4703-9220 |
| authorships[2].author.display_name | Libo Jiang |
| authorships[2].countries | CN |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[2].affiliations[0].raw_affiliation_string | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[2].affiliations[1].institution_ids | https://openalex.org/I31683504 |
| authorships[2].affiliations[1].raw_affiliation_string | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[2].institutions[0].id | https://openalex.org/I31683504 |
| authorships[2].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[2].institutions[0].country_code | CN |
| authorships[2].institutions[0].display_name | Beijing Forestry University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Libo Jiang |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[3].author.id | https://openalex.org/A5008729081 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Meixia Ye |
| authorships[3].countries | CN |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[3].affiliations[0].raw_affiliation_string | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[3].affiliations[1].institution_ids | https://openalex.org/I31683504 |
| authorships[3].affiliations[1].raw_affiliation_string | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[3].institutions[0].id | https://openalex.org/I31683504 |
| authorships[3].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[3].institutions[0].country_code | CN |
| authorships[3].institutions[0].display_name | Beijing Forestry University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Meixia Ye |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[4].author.id | https://openalex.org/A5072773597 |
| authorships[4].author.orcid | https://orcid.org/0000-0001-9098-9980 |
| authorships[4].author.display_name | Xiaoqing He |
| authorships[4].countries | CN |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[4].affiliations[0].raw_affiliation_string | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[4].affiliations[1].institution_ids | https://openalex.org/I31683504 |
| authorships[4].affiliations[1].raw_affiliation_string | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[4].institutions[0].id | https://openalex.org/I31683504 |
| authorships[4].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[4].institutions[0].country_code | CN |
| authorships[4].institutions[0].display_name | Beijing Forestry University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Xiaoqing He |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[5].author.id | https://openalex.org/A5032833513 |
| authorships[5].author.orcid | https://orcid.org/0000-0001-6412-0835 |
| authorships[5].author.display_name | Yi Jin |
| authorships[5].countries | CN |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[5].affiliations[0].raw_affiliation_string | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[5].affiliations[1].institution_ids | https://openalex.org/I31683504 |
| authorships[5].affiliations[1].raw_affiliation_string | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[5].institutions[0].id | https://openalex.org/I31683504 |
| authorships[5].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[5].institutions[0].country_code | CN |
| authorships[5].institutions[0].display_name | Beijing Forestry University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Yi Jin |
| authorships[5].is_corresponding | True |
| authorships[5].raw_affiliation_strings | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[6].author.id | https://openalex.org/A5031422180 |
| authorships[6].author.orcid | https://orcid.org/0000-0001-8350-7779 |
| authorships[6].author.display_name | Rongling Wu |
| authorships[6].countries | CN, US |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I31683504 |
| authorships[6].affiliations[0].raw_affiliation_string | College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[6].affiliations[1].institution_ids | https://openalex.org/I31683504 |
| authorships[6].affiliations[1].raw_affiliation_string | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China |
| authorships[6].affiliations[2].institution_ids | https://openalex.org/I130769515 |
| authorships[6].affiliations[2].raw_affiliation_string | Department of Public Health Sciences and Statistics, Center for Statistical Genetics, The Pennsylvania State University, Hershey, PA, United States |
| authorships[6].institutions[0].id | https://openalex.org/I31683504 |
| authorships[6].institutions[0].ror | https://ror.org/04xv2pc41 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I1327237609, https://openalex.org/I31683504, https://openalex.org/I4210127390 |
| authorships[6].institutions[0].country_code | CN |
| authorships[6].institutions[0].display_name | Beijing Forestry University |
| authorships[6].institutions[1].id | https://openalex.org/I130769515 |
| authorships[6].institutions[1].ror | https://ror.org/04p491231 |
| authorships[6].institutions[1].type | education |
| authorships[6].institutions[1].lineage | https://openalex.org/I130769515 |
| authorships[6].institutions[1].country_code | US |
| authorships[6].institutions[1].display_name | Pennsylvania State University |
| authorships[6].author_position | last |
| authorships[6].raw_author_name | Rongling Wu |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Center for Computational Biology, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China, Department of Public Health Sciences and Statistics, Center for Statistical Genetics, The Pennsylvania State University, Hershey, PA, United States |
| has_content.pdf | True |
| has_content.grobid_xml | True |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdf |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Functional Mapping of Phenotypic Plasticity of Staphylococcus aureus Under Vancomycin Pressure |
| has_fulltext | True |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10195 |
| primary_topic.field.id | https://openalex.org/fields/27 |
| primary_topic.field.display_name | Medicine |
| primary_topic.score | 0.9970999956130981 |
| primary_topic.domain.id | https://openalex.org/domains/4 |
| primary_topic.domain.display_name | Health Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2725 |
| primary_topic.subfield.display_name | Infectious Diseases |
| primary_topic.display_name | Antimicrobial Resistance in Staphylococcus |
| related_works | https://openalex.org/W4283582241, https://openalex.org/W3098014387, https://openalex.org/W2754869062, https://openalex.org/W2986919234, https://openalex.org/W2982417329, https://openalex.org/W3195469724, https://openalex.org/W2923859939, https://openalex.org/W2142550298, https://openalex.org/W2148749248, https://openalex.org/W1969370271 |
| cited_by_count | 5 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 2 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| counts_by_year[2].year | 2022 |
| counts_by_year[2].cited_by_count | 1 |
| locations_count | 5 |
| best_oa_location.id | doi:10.3389/fmicb.2021.696730 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2484704948 |
| best_oa_location.source.issn | 1664-302X |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 1664-302X |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Frontiers in Microbiology |
| best_oa_location.source.host_organization | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_name | Frontiers Media |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| best_oa_location.source.host_organization_lineage_names | Frontiers Media |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdf |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Frontiers in Microbiology |
| best_oa_location.landing_page_url | https://doi.org/10.3389/fmicb.2021.696730 |
| primary_location.id | doi:10.3389/fmicb.2021.696730 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2484704948 |
| primary_location.source.issn | 1664-302X |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 1664-302X |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Frontiers in Microbiology |
| primary_location.source.host_organization | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_name | Frontiers Media |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320527 |
| primary_location.source.host_organization_lineage_names | Frontiers Media |
| primary_location.license | cc-by |
| primary_location.pdf_url | https://www.frontiersin.org/articles/10.3389/fmicb.2021.696730/pdf |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Frontiers in Microbiology |
| primary_location.landing_page_url | https://doi.org/10.3389/fmicb.2021.696730 |
| publication_date | 2021-09-09 |
| publication_year | 2021 |
| referenced_works | https://openalex.org/W2131843590, https://openalex.org/W2079689510, https://openalex.org/W2139704494, https://openalex.org/W2521143611, https://openalex.org/W1987317996, https://openalex.org/W3002978601, https://openalex.org/W3097971644, https://openalex.org/W3121491346, https://openalex.org/W1991784736, https://openalex.org/W2914706176, https://openalex.org/W2107289345, https://openalex.org/W2730206157, https://openalex.org/W2111197995, https://openalex.org/W2125860227, https://openalex.org/W2110055201, https://openalex.org/W2948444604, https://openalex.org/W3006289601, https://openalex.org/W1983784984, https://openalex.org/W2768143668, https://openalex.org/W2020962641, https://openalex.org/W2531317781, https://openalex.org/W2157607613, https://openalex.org/W2903590625, https://openalex.org/W2168683060, https://openalex.org/W2913252238, https://openalex.org/W2113784410, https://openalex.org/W2898111315, https://openalex.org/W1789064953, https://openalex.org/W2108234281, https://openalex.org/W1967731019, https://openalex.org/W2116901749, https://openalex.org/W2898974435, https://openalex.org/W1944381374, https://openalex.org/W3047696772, https://openalex.org/W2953340095, https://openalex.org/W3081666419, https://openalex.org/W2304212006, https://openalex.org/W2155782878, https://openalex.org/W2138556224, https://openalex.org/W2155476515, https://openalex.org/W2939431273, https://openalex.org/W2014466662, https://openalex.org/W2535369069, https://openalex.org/W3038123490, https://openalex.org/W3030441856, https://openalex.org/W2510547218, https://openalex.org/W3126899922, https://openalex.org/W2914002890, https://openalex.org/W2571097790, https://openalex.org/W1546892755, https://openalex.org/W1996952375, https://openalex.org/W2050180108, https://openalex.org/W2034637902, https://openalex.org/W3023719018, https://openalex.org/W3118686772 |
| referenced_works_count | 55 |
| abstract_inverted_index.( | 151, 156 |
| abstract_inverted_index.6 | 79 |
| abstract_inverted_index.a | 11, 55, 82, 89, 135, 179 |
| abstract_inverted_index.), | 153, 158 |
| abstract_inverted_index.2, | 76 |
| abstract_inverted_index.4, | 77 |
| abstract_inverted_index.78 | 114 |
| abstract_inverted_index.In | 48 |
| abstract_inverted_index.S. | 111, 168, 188 |
| abstract_inverted_index.We | 85 |
| abstract_inverted_index.by | 10, 26 |
| abstract_inverted_index.in | 14, 34, 81, 138, 201 |
| abstract_inverted_index.is | 2 |
| abstract_inverted_index.of | 5, 45, 64, 73, 107, 109, 123, 145, 187, 198, 204 |
| abstract_inverted_index.to | 16, 21, 23, 60, 70, 98, 142, 166 |
| abstract_inverted_index.we | 37, 51 |
| abstract_inverted_index.(0, | 75 |
| abstract_inverted_index.Our | 173 |
| abstract_inverted_index.and | 29, 53, 78, 127, 130, 162, 196 |
| abstract_inverted_index.for | 92, 181 |
| abstract_inverted_index.how | 65 |
| abstract_inverted_index.its | 32 |
| abstract_inverted_index.map | 99 |
| abstract_inverted_index.the | 3, 42, 104, 124, 143, 183, 194, 202 |
| abstract_inverted_index.gene | 175 |
| abstract_inverted_index.help | 167 |
| abstract_inverted_index.know | 39 |
| abstract_inverted_index.loci | 101 |
| abstract_inverted_index.play | 134 |
| abstract_inverted_index.role | 137 |
| abstract_inverted_index.sdrC | 157 |
| abstract_inverted_index.that | 102 |
| abstract_inverted_index.this | 49 |
| abstract_inverted_index.time | 83 |
| abstract_inverted_index.tool | 180 |
| abstract_inverted_index.walR | 152 |
| abstract_inverted_index.were | 119, 164 |
| abstract_inverted_index.These | 147 |
| abstract_inverted_index.about | 41 |
| abstract_inverted_index.adapt | 22 |
| abstract_inverted_index.data, | 97 |
| abstract_inverted_index.found | 165 |
| abstract_inverted_index.genes | 132 |
| abstract_inverted_index.might | 133 |
| abstract_inverted_index.model | 91 |
| abstract_inverted_index.seven | 131, 148 |
| abstract_inverted_index.still | 38 |
| abstract_inverted_index.study | 58, 203 |
| abstract_inverted_index.under | 190 |
| abstract_inverted_index.using | 95 |
| abstract_inverted_index.whole | 125 |
| abstract_inverted_index.(GWAS) | 59 |
| abstract_inverted_index.aureus | 67, 112, 169, 189 |
| abstract_inverted_index.genes, | 149 |
| abstract_inverted_index.growth | 28, 105, 126 |
| abstract_inverted_index.little | 40 |
| abstract_inverted_index.reveal | 61 |
| abstract_inverted_index.single | 12, 116 |
| abstract_inverted_index.study, | 50 |
| abstract_inverted_index.traits | 8 |
| abstract_inverted_index.Despite | 31 |
| abstract_inverted_index.changes | 25 |
| abstract_inverted_index.control | 44 |
| abstract_inverted_index.course. | 84 |
| abstract_inverted_index.dynamic | 90, 174 |
| abstract_inverted_index.genetic | 43, 62, 93 |
| abstract_inverted_index.mapping | 94, 176, 200 |
| abstract_inverted_index.mediate | 103 |
| abstract_inverted_index.pivotal | 136 |
| abstract_inverted_index.respond | 69 |
| abstract_inverted_index.strains | 68 |
| abstract_inverted_index.various | 6 |
| abstract_inverted_index.μg/mL) | 80 |
| abstract_inverted_index.analysis | 122 |
| abstract_inverted_index.changes, | 18 |
| abstract_inverted_index.designed | 52 |
| abstract_inverted_index.enabling | 19 |
| abstract_inverted_index.genotype | 13 |
| abstract_inverted_index.mapping, | 88 |
| abstract_inverted_index.pressure | 144 |
| abstract_inverted_index.process, | 129 |
| abstract_inverted_index.produced | 9 |
| abstract_inverted_index.provides | 178 |
| abstract_inverted_index.regulate | 170 |
| abstract_inverted_index.response | 15 |
| abstract_inverted_index.specific | 100 |
| abstract_inverted_index.strains. | 113 |
| abstract_inverted_index.studies, | 36 |
| abstract_inverted_index.abundance | 108 |
| abstract_inverted_index.bacterial | 205 |
| abstract_inverted_index.conducted | 54 |
| abstract_inverted_index.following | 121 |
| abstract_inverted_index.governing | 139 |
| abstract_inverted_index.organisms | 20 |
| abstract_inverted_index.potential | 197 |
| abstract_inverted_index.pressure, | 192 |
| abstract_inverted_index.pressure. | 172 |
| abstract_inverted_index.technique | 177 |
| abstract_inverted_index.Phenotypic | 0 |
| abstract_inverted_index.antibiotic | 171 |
| abstract_inverted_index.dissecting | 182 |
| abstract_inverted_index.exhibition | 4 |
| abstract_inverted_index.functional | 87, 199 |
| abstract_inverted_index.identified | 120 |
| abstract_inverted_index.increasing | 71 |
| abstract_inverted_index.mechanisms | 186 |
| abstract_inverted_index.nucleotide | 117 |
| abstract_inverted_index.phenotypic | 7, 46, 140, 184, 206 |
| abstract_inverted_index.plasticity | 1, 141, 185 |
| abstract_inverted_index.vancomycin | 74, 191 |
| abstract_inverted_index.association | 57 |
| abstract_inverted_index.development | 128 |
| abstract_inverted_index.emphasizing | 193 |
| abstract_inverted_index.feasibility | 195 |
| abstract_inverted_index.genome-wide | 56 |
| abstract_inverted_index.implemented | 86 |
| abstract_inverted_index.maintaining | 27 |
| abstract_inverted_index.plasticity. | 47, 207 |
| abstract_inverted_index.significant | 115 |
| abstract_inverted_index.vancomycin. | 146 |
| abstract_inverted_index.architecture | 63 |
| abstract_inverted_index.evolutionary | 35 |
| abstract_inverted_index.longitudinal | 96 |
| abstract_inverted_index.significance | 33 |
| abstract_inverted_index.trajectories | 106 |
| abstract_inverted_index.SAOUHSC_00020 | 150 |
| abstract_inverted_index.SAOUHSC_00544 | 155 |
| abstract_inverted_index.environmental | 17, 24 |
| abstract_inverted_index.polymorphisms | 118 |
| abstract_inverted_index.reproduction. | 30 |
| abstract_inverted_index.SAOUHSC_00025, | 160 |
| abstract_inverted_index.SAOUHSC_00169, | 161 |
| abstract_inverted_index.SAOUHSC_00176, | 154 |
| abstract_inverted_index.SAOUHSC_02023, | 163 |
| abstract_inverted_index.SAOUHSC_02998, | 159 |
| abstract_inverted_index.Staphylococcus | 66 |
| abstract_inverted_index.concentrations | 72 |
| abstract_inverted_index.vancomycin-exposed | 110 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 89 |
| corresponding_author_ids | https://openalex.org/A5032833513 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 7 |
| corresponding_institution_ids | https://openalex.org/I31683504 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/15 |
| sustainable_development_goals[0].score | 0.6399999856948853 |
| sustainable_development_goals[0].display_name | Life in Land |
| citation_normalized_percentile.value | 0.54245439 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |