Genetic Structure and Diversity Study of Cassava (<i>Manihot esculenta</i>) Germplasm for African Cassava Mosaic Disease and Fresh Storage Root Yield Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.4236/ojgen.2023.131002
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.4236/ojgen.2023.131002
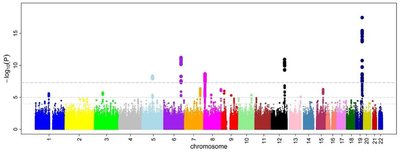
A better understanding of population structure and genetic diversity among cassava germplasm for African cassava mosaic disease and fresh root yield traits is useful for cassava improvement programme. Phenotype-based selection for these traits is cumbersome due to phenotypic plasticity and difficulty in screening of phenotypic-induced variations. This study assessed quantitative trait loci (QTL) regions associated with African cassava mosaic disease (ACMD) and fresh storage root yield (FSRY) in 131 cassava (Manihot esculenta) genotypes using a genome-wide association study (GWAS). The single nucleotide polymorphism (SNP) loci and associated candidate genes, when validated, would be a valuable resource for marker-assisted selection in the breeding process for development of new cassava genotypes with improved resistance to ACMD and desirable high root yield. Population structure analysis using 12,500 SNPs differentiated the 131 genotypes into five distinct sub-groups (K = 5). Marker-trait association (MTA) analysis using the generalized linear model identified two QTL regions significant for ACMD and three for FSRY. This study demonstrated that DArTseq markers are useful genomic resources for genome-wide association studies of ACMD and FSRY traits in cassava for the acceleration of varietal development and release.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.4236/ojgen.2023.131002
- OA Status
- diamond
- Cited By
- 7
- References
- 50
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4324013754
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4324013754Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.4236/ojgen.2023.131002Digital Object Identifier
- Title
-
Genetic Structure and Diversity Study of Cassava (<i>Manihot esculenta</i>) Germplasm for African Cassava Mosaic Disease and Fresh Storage Root YieldWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-01-01Full publication date if available
- Authors
-
Janatu Veronica Sesay, Aiah Lebbie, Richard Wadsworth, Ephraim Nuwamanya, Souleymane Bado, Prince Emmanuel NormanList of authors in order
- Landing page
-
https://doi.org/10.4236/ojgen.2023.131002Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
diamondOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.4236/ojgen.2023.131002Direct OA link when available
- Concepts
-
Germplasm, Biology, Quantitative trait locus, Single-nucleotide polymorphism, Genome-wide association study, Genetic diversity, Population, Trait, Genotype, Genetic association, Genetics, Biotechnology, Agronomy, Gene, Medicine, Environmental health, Programming language, Computer scienceTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
7Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 4, 2024: 3Per-year citation counts (last 5 years)
- References (count)
-
50Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4324013754 |
|---|---|
| doi | https://doi.org/10.4236/ojgen.2023.131002 |
| ids.doi | https://doi.org/10.4236/ojgen.2023.131002 |
| ids.openalex | https://openalex.org/W4324013754 |
| fwci | 1.84926163 |
| type | article |
| title | Genetic Structure and Diversity Study of Cassava (<i>Manihot esculenta</i>) Germplasm for African Cassava Mosaic Disease and Fresh Storage Root Yield |
| biblio.issue | 01 |
| biblio.volume | 13 |
| biblio.last_page | 47 |
| biblio.first_page | 23 |
| topics[0].id | https://openalex.org/T12305 |
| topics[0].field.id | https://openalex.org/fields/11 |
| topics[0].field.display_name | Agricultural and Biological Sciences |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1110 |
| topics[0].subfield.display_name | Plant Science |
| topics[0].display_name | Cassava research and cyanide |
| topics[1].id | https://openalex.org/T12115 |
| topics[1].field.id | https://openalex.org/fields/11 |
| topics[1].field.display_name | Agricultural and Biological Sciences |
| topics[1].score | 0.9936000108718872 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1110 |
| topics[1].subfield.display_name | Plant Science |
| topics[1].display_name | Genetic and Environmental Crop Studies |
| topics[2].id | https://openalex.org/T12795 |
| topics[2].field.id | https://openalex.org/fields/11 |
| topics[2].field.display_name | Agricultural and Biological Sciences |
| topics[2].score | 0.9907000064849854 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1110 |
| topics[2].subfield.display_name | Plant Science |
| topics[2].display_name | Banana Cultivation and Research |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C2777461220 |
| concepts[0].level | 2 |
| concepts[0].score | 0.8894615769386292 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q589029 |
| concepts[0].display_name | Germplasm |
| concepts[1].id | https://openalex.org/C86803240 |
| concepts[1].level | 0 |
| concepts[1].score | 0.8428187966346741 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[1].display_name | Biology |
| concepts[2].id | https://openalex.org/C81941488 |
| concepts[2].level | 3 |
| concepts[2].score | 0.6011994481086731 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q853421 |
| concepts[2].display_name | Quantitative trait locus |
| concepts[3].id | https://openalex.org/C153209595 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5664167404174805 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[3].display_name | Single-nucleotide polymorphism |
| concepts[4].id | https://openalex.org/C106208931 |
| concepts[4].level | 5 |
| concepts[4].score | 0.5259506702423096 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[4].display_name | Genome-wide association study |
| concepts[5].id | https://openalex.org/C81977670 |
| concepts[5].level | 3 |
| concepts[5].score | 0.5135212540626526 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q585259 |
| concepts[5].display_name | Genetic diversity |
| concepts[6].id | https://openalex.org/C2908647359 |
| concepts[6].level | 2 |
| concepts[6].score | 0.5093973278999329 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q2625603 |
| concepts[6].display_name | Population |
| concepts[7].id | https://openalex.org/C106934330 |
| concepts[7].level | 2 |
| concepts[7].score | 0.4656297564506531 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1971873 |
| concepts[7].display_name | Trait |
| concepts[8].id | https://openalex.org/C135763542 |
| concepts[8].level | 3 |
| concepts[8].score | 0.4586619734764099 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[8].display_name | Genotype |
| concepts[9].id | https://openalex.org/C186413461 |
| concepts[9].level | 5 |
| concepts[9].score | 0.44757890701293945 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q744727 |
| concepts[9].display_name | Genetic association |
| concepts[10].id | https://openalex.org/C54355233 |
| concepts[10].level | 1 |
| concepts[10].score | 0.43420472741127014 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[10].display_name | Genetics |
| concepts[11].id | https://openalex.org/C150903083 |
| concepts[11].level | 1 |
| concepts[11].score | 0.4094831943511963 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q7108 |
| concepts[11].display_name | Biotechnology |
| concepts[12].id | https://openalex.org/C6557445 |
| concepts[12].level | 1 |
| concepts[12].score | 0.34225308895111084 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q173113 |
| concepts[12].display_name | Agronomy |
| concepts[13].id | https://openalex.org/C104317684 |
| concepts[13].level | 2 |
| concepts[13].score | 0.21726223826408386 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[13].display_name | Gene |
| concepts[14].id | https://openalex.org/C71924100 |
| concepts[14].level | 0 |
| concepts[14].score | 0.06304967403411865 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[14].display_name | Medicine |
| concepts[15].id | https://openalex.org/C99454951 |
| concepts[15].level | 1 |
| concepts[15].score | 0.0 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q932068 |
| concepts[15].display_name | Environmental health |
| concepts[16].id | https://openalex.org/C199360897 |
| concepts[16].level | 1 |
| concepts[16].score | 0.0 |
| concepts[16].wikidata | https://www.wikidata.org/wiki/Q9143 |
| concepts[16].display_name | Programming language |
| concepts[17].id | https://openalex.org/C41008148 |
| concepts[17].level | 0 |
| concepts[17].score | 0.0 |
| concepts[17].wikidata | https://www.wikidata.org/wiki/Q21198 |
| concepts[17].display_name | Computer science |
| keywords[0].id | https://openalex.org/keywords/germplasm |
| keywords[0].score | 0.8894615769386292 |
| keywords[0].display_name | Germplasm |
| keywords[1].id | https://openalex.org/keywords/biology |
| keywords[1].score | 0.8428187966346741 |
| keywords[1].display_name | Biology |
| keywords[2].id | https://openalex.org/keywords/quantitative-trait-locus |
| keywords[2].score | 0.6011994481086731 |
| keywords[2].display_name | Quantitative trait locus |
| keywords[3].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[3].score | 0.5664167404174805 |
| keywords[3].display_name | Single-nucleotide polymorphism |
| keywords[4].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[4].score | 0.5259506702423096 |
| keywords[4].display_name | Genome-wide association study |
| keywords[5].id | https://openalex.org/keywords/genetic-diversity |
| keywords[5].score | 0.5135212540626526 |
| keywords[5].display_name | Genetic diversity |
| keywords[6].id | https://openalex.org/keywords/population |
| keywords[6].score | 0.5093973278999329 |
| keywords[6].display_name | Population |
| keywords[7].id | https://openalex.org/keywords/trait |
| keywords[7].score | 0.4656297564506531 |
| keywords[7].display_name | Trait |
| keywords[8].id | https://openalex.org/keywords/genotype |
| keywords[8].score | 0.4586619734764099 |
| keywords[8].display_name | Genotype |
| keywords[9].id | https://openalex.org/keywords/genetic-association |
| keywords[9].score | 0.44757890701293945 |
| keywords[9].display_name | Genetic association |
| keywords[10].id | https://openalex.org/keywords/genetics |
| keywords[10].score | 0.43420472741127014 |
| keywords[10].display_name | Genetics |
| keywords[11].id | https://openalex.org/keywords/biotechnology |
| keywords[11].score | 0.4094831943511963 |
| keywords[11].display_name | Biotechnology |
| keywords[12].id | https://openalex.org/keywords/agronomy |
| keywords[12].score | 0.34225308895111084 |
| keywords[12].display_name | Agronomy |
| keywords[13].id | https://openalex.org/keywords/gene |
| keywords[13].score | 0.21726223826408386 |
| keywords[13].display_name | Gene |
| keywords[14].id | https://openalex.org/keywords/medicine |
| keywords[14].score | 0.06304967403411865 |
| keywords[14].display_name | Medicine |
| language | en |
| locations[0].id | doi:10.4236/ojgen.2023.131002 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S2765021627 |
| locations[0].source.issn | 2162-4453, 2162-4461 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2162-4453 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | Open Journal of Genetics |
| locations[0].source.host_organization | https://openalex.org/P4310320480 |
| locations[0].source.host_organization_name | Scientific Research Publishing |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320480 |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Open Journal of Genetics |
| locations[0].landing_page_url | https://doi.org/10.4236/ojgen.2023.131002 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5077458830 |
| authorships[0].author.orcid | |
| authorships[0].author.display_name | Janatu Veronica Sesay |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Janatu Veronica Sesay |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5069645163 |
| authorships[1].author.orcid | https://orcid.org/0000-0001-8725-3593 |
| authorships[1].author.display_name | Aiah Lebbie |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Aiah Lebbie |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5008812095 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-0593-8826 |
| authorships[2].author.display_name | Richard Wadsworth |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Richard Wadsworth |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5028310092 |
| authorships[3].author.orcid | https://orcid.org/0000-0001-8925-0628 |
| authorships[3].author.display_name | Ephraim Nuwamanya |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Ephraim Nuwamanya |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5043488486 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-1413-2517 |
| authorships[4].author.display_name | Souleymane Bado |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Souleymane Bado |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5065982491 |
| authorships[5].author.orcid | https://orcid.org/0000-0002-0150-8610 |
| authorships[5].author.display_name | Prince Emmanuel Norman |
| authorships[5].author_position | last |
| authorships[5].raw_author_name | Prince Emmanuel Norman |
| authorships[5].is_corresponding | False |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.4236/ojgen.2023.131002 |
| open_access.oa_status | diamond |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Genetic Structure and Diversity Study of Cassava (<i>Manihot esculenta</i>) Germplasm for African Cassava Mosaic Disease and Fresh Storage Root Yield |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T12305 |
| primary_topic.field.id | https://openalex.org/fields/11 |
| primary_topic.field.display_name | Agricultural and Biological Sciences |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1110 |
| primary_topic.subfield.display_name | Plant Science |
| primary_topic.display_name | Cassava research and cyanide |
| related_works | https://openalex.org/W4231021975, https://openalex.org/W2176884741, https://openalex.org/W2734576438, https://openalex.org/W2587112589, https://openalex.org/W2743853223, https://openalex.org/W3082508018, https://openalex.org/W2884987722, https://openalex.org/W4385987399, https://openalex.org/W4319077693, https://openalex.org/W4311569799 |
| cited_by_count | 7 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 4 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 3 |
| locations_count | 1 |
| best_oa_location.id | doi:10.4236/ojgen.2023.131002 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S2765021627 |
| best_oa_location.source.issn | 2162-4453, 2162-4461 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2162-4453 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | Open Journal of Genetics |
| best_oa_location.source.host_organization | https://openalex.org/P4310320480 |
| best_oa_location.source.host_organization_name | Scientific Research Publishing |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320480 |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Open Journal of Genetics |
| best_oa_location.landing_page_url | https://doi.org/10.4236/ojgen.2023.131002 |
| primary_location.id | doi:10.4236/ojgen.2023.131002 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S2765021627 |
| primary_location.source.issn | 2162-4453, 2162-4461 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2162-4453 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | Open Journal of Genetics |
| primary_location.source.host_organization | https://openalex.org/P4310320480 |
| primary_location.source.host_organization_name | Scientific Research Publishing |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320480 |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Open Journal of Genetics |
| primary_location.landing_page_url | https://doi.org/10.4236/ojgen.2023.131002 |
| publication_date | 2023-01-01 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2089607238, https://openalex.org/W2071756433, https://openalex.org/W2005419745, https://openalex.org/W2151291924, https://openalex.org/W2166204226, https://openalex.org/W2746908215, https://openalex.org/W2484746334, https://openalex.org/W2137674766, https://openalex.org/W3004379071, https://openalex.org/W2073970020, https://openalex.org/W3181461972, https://openalex.org/W2947553484, https://openalex.org/W2161238842, https://openalex.org/W2206346562, https://openalex.org/W2032923546, https://openalex.org/W2029181991, https://openalex.org/W592897681, https://openalex.org/W2810102070, https://openalex.org/W2474436508, https://openalex.org/W2080737684, https://openalex.org/W1546350498, https://openalex.org/W1993849755, https://openalex.org/W2486415217, https://openalex.org/W2801987717, https://openalex.org/W2006807779, https://openalex.org/W1971003087, https://openalex.org/W2335517459, https://openalex.org/W2149992227, https://openalex.org/W2161633633, https://openalex.org/W2153947402, https://openalex.org/W2098126593, https://openalex.org/W2158489424, https://openalex.org/W2161339576, https://openalex.org/W2035839217, https://openalex.org/W2883251903, https://openalex.org/W2951328066, https://openalex.org/W3046908823, https://openalex.org/W2003073123, https://openalex.org/W3016476173, https://openalex.org/W2791978259, https://openalex.org/W2090075367, https://openalex.org/W2883759060, https://openalex.org/W2804103085, https://openalex.org/W2949299396, https://openalex.org/W2994597697, https://openalex.org/W2082899584, https://openalex.org/W3017977577, https://openalex.org/W2122138951, https://openalex.org/W2797088263, https://openalex.org/W2062791755 |
| referenced_works_count | 50 |
| abstract_inverted_index.= | 134 |
| abstract_inverted_index.A | 0 |
| abstract_inverted_index.a | 74, 93 |
| abstract_inverted_index.(K | 133 |
| abstract_inverted_index.be | 92 |
| abstract_inverted_index.in | 41, 67, 99, 175 |
| abstract_inverted_index.is | 22, 33 |
| abstract_inverted_index.of | 3, 43, 105, 170, 180 |
| abstract_inverted_index.to | 36, 112 |
| abstract_inverted_index.131 | 68, 127 |
| abstract_inverted_index.5). | 135 |
| abstract_inverted_index.QTL | 147 |
| abstract_inverted_index.The | 79 |
| abstract_inverted_index.and | 6, 17, 39, 61, 85, 114, 152, 172, 183 |
| abstract_inverted_index.are | 162 |
| abstract_inverted_index.due | 35 |
| abstract_inverted_index.for | 12, 24, 30, 96, 103, 150, 154, 166, 177 |
| abstract_inverted_index.new | 106 |
| abstract_inverted_index.the | 100, 126, 141, 178 |
| abstract_inverted_index.two | 146 |
| abstract_inverted_index.ACMD | 113, 151, 171 |
| abstract_inverted_index.FSRY | 173 |
| abstract_inverted_index.SNPs | 124 |
| abstract_inverted_index.This | 46, 156 |
| abstract_inverted_index.five | 130 |
| abstract_inverted_index.high | 116 |
| abstract_inverted_index.into | 129 |
| abstract_inverted_index.loci | 51, 84 |
| abstract_inverted_index.root | 19, 64, 117 |
| abstract_inverted_index.that | 159 |
| abstract_inverted_index.when | 89 |
| abstract_inverted_index.with | 55, 109 |
| abstract_inverted_index.(MTA) | 138 |
| abstract_inverted_index.(QTL) | 52 |
| abstract_inverted_index.(SNP) | 83 |
| abstract_inverted_index.FSRY. | 155 |
| abstract_inverted_index.among | 9 |
| abstract_inverted_index.fresh | 18, 62 |
| abstract_inverted_index.model | 144 |
| abstract_inverted_index.study | 47, 77, 157 |
| abstract_inverted_index.these | 31 |
| abstract_inverted_index.three | 153 |
| abstract_inverted_index.trait | 50 |
| abstract_inverted_index.using | 73, 122, 140 |
| abstract_inverted_index.would | 91 |
| abstract_inverted_index.yield | 20, 65 |
| abstract_inverted_index.(ACMD) | 60 |
| abstract_inverted_index.(FSRY) | 66 |
| abstract_inverted_index.12,500 | 123 |
| abstract_inverted_index.better | 1 |
| abstract_inverted_index.genes, | 88 |
| abstract_inverted_index.linear | 143 |
| abstract_inverted_index.mosaic | 15, 58 |
| abstract_inverted_index.single | 80 |
| abstract_inverted_index.traits | 21, 32, 174 |
| abstract_inverted_index.useful | 23, 163 |
| abstract_inverted_index.yield. | 118 |
| abstract_inverted_index.(GWAS). | 78 |
| abstract_inverted_index.African | 13, 56 |
| abstract_inverted_index.DArTseq | 160 |
| abstract_inverted_index.cassava | 10, 14, 25, 57, 69, 107, 176 |
| abstract_inverted_index.disease | 16, 59 |
| abstract_inverted_index.genetic | 7 |
| abstract_inverted_index.genomic | 164 |
| abstract_inverted_index.markers | 161 |
| abstract_inverted_index.process | 102 |
| abstract_inverted_index.regions | 53, 148 |
| abstract_inverted_index.storage | 63 |
| abstract_inverted_index.studies | 169 |
| abstract_inverted_index.(Manihot | 70 |
| abstract_inverted_index.analysis | 121, 139 |
| abstract_inverted_index.assessed | 48 |
| abstract_inverted_index.breeding | 101 |
| abstract_inverted_index.distinct | 131 |
| abstract_inverted_index.improved | 110 |
| abstract_inverted_index.release. | 184 |
| abstract_inverted_index.resource | 95 |
| abstract_inverted_index.valuable | 94 |
| abstract_inverted_index.varietal | 181 |
| abstract_inverted_index.candidate | 87 |
| abstract_inverted_index.desirable | 115 |
| abstract_inverted_index.diversity | 8 |
| abstract_inverted_index.genotypes | 72, 108, 128 |
| abstract_inverted_index.germplasm | 11 |
| abstract_inverted_index.resources | 165 |
| abstract_inverted_index.screening | 42 |
| abstract_inverted_index.selection | 29, 98 |
| abstract_inverted_index.structure | 5, 120 |
| abstract_inverted_index.Population | 119 |
| abstract_inverted_index.associated | 54, 86 |
| abstract_inverted_index.cumbersome | 34 |
| abstract_inverted_index.difficulty | 40 |
| abstract_inverted_index.esculenta) | 71 |
| abstract_inverted_index.identified | 145 |
| abstract_inverted_index.nucleotide | 81 |
| abstract_inverted_index.phenotypic | 37 |
| abstract_inverted_index.plasticity | 38 |
| abstract_inverted_index.population | 4 |
| abstract_inverted_index.programme. | 27 |
| abstract_inverted_index.resistance | 111 |
| abstract_inverted_index.sub-groups | 132 |
| abstract_inverted_index.validated, | 90 |
| abstract_inverted_index.association | 76, 137, 168 |
| abstract_inverted_index.development | 104, 182 |
| abstract_inverted_index.generalized | 142 |
| abstract_inverted_index.genome-wide | 75, 167 |
| abstract_inverted_index.improvement | 26 |
| abstract_inverted_index.significant | 149 |
| abstract_inverted_index.variations. | 45 |
| abstract_inverted_index.Marker-trait | 136 |
| abstract_inverted_index.acceleration | 179 |
| abstract_inverted_index.demonstrated | 158 |
| abstract_inverted_index.polymorphism | 82 |
| abstract_inverted_index.quantitative | 49 |
| abstract_inverted_index.understanding | 2 |
| abstract_inverted_index.differentiated | 125 |
| abstract_inverted_index.Phenotype-based | 28 |
| abstract_inverted_index.marker-assisted | 97 |
| abstract_inverted_index.phenotypic-induced | 44 |
| cited_by_percentile_year.max | 98 |
| cited_by_percentile_year.min | 96 |
| countries_distinct_count | 0 |
| institutions_distinct_count | 6 |
| citation_normalized_percentile.value | 0.91105633 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |