Genetic variants linked to the phenotypic outcome of invasive disease and carriage of Neisseria meningitidis Article Swipe
 YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1099/mgen.0.001124
YOU?
·
· 2023
· Open Access
·
· DOI: https://doi.org/10.1099/mgen.0.001124
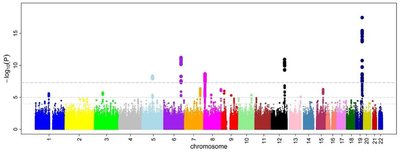
Neisseria meningitidis can be a human commensal in the upper respiratory tract but is also capable of causing invasive diseases such as meningococcal meningitis and septicaemia. No specific genetic markers have been detected to distinguish carriage from disease isolates. The aim here was to find genetic traits that could be linked to phenotypic outcomes associated with carriage versus invasive N. meningitidis disease through a bacterial genome-wide association study (GWAS). In this study, invasive N. meningitidis isolates collected in Sweden ( n =103) and carriage isolates collected at Örebro University, Sweden ( n =213) 2018–2019 were analysed. The GWAS analysis, treeWAS , was applied to single-nucleotide polymorphisms (SNPs), genes and k-mers. One gene and one non-synonymous SNP were associated with invasive disease and seven genes and one non-synonymous SNP were associated with carriage isolates. The gene associated with invasive disease encodes a phage transposase (NEIS1048), and the associated invasive SNP glmU S373C encodes the enzyme N-acetylglucosamine 1-phosphate (GlcNAC 1-P) uridyltransferase. Of the genes associated with carriage isolates, a gene variant of porB encoding PorB class 3, the genes pilE / pilS and tspB have known functions. The SNP associated with carriage was fkbp D33N, encoding a FK506-binding protein (FKBP). K-mers from PilS , tbpB and tspB were found to be associated with carriage, while k-mers from mtrD and tbpA were associated with invasiveness. In the genes fkbp , glmU , PilC and pilE , k-mers were found that were associated with both carriage and invasive isolates, indicating that specific variations within these genes could play a role in invasiveness. The data presented here highlight genetic traits that are significantly associated with invasive or carriage N. meningitidis across the species population. These traits could prove essential to our understanding of the pathogenicity of N. meningitidis and could help to identify future vaccine targets.
Related Topics
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1099/mgen.0.001124
- OA Status
- gold
- Cited By
- 5
- References
- 75
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4387893260
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4387893260Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1099/mgen.0.001124Digital Object Identifier
- Title
-
Genetic variants linked to the phenotypic outcome of invasive disease and carriage of Neisseria meningitidisWork title
- Type
-
articleOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2023Year of publication
- Publication date
-
2023-10-24Full publication date if available
- Authors
-
Lorraine Eriksson, Thor Bech Johannesen, Bianca Stenmark, Susanne Jacobsson, Olof Säll, Sara Thulin Hedberg, Hans Fredlund, Marc Stegger, Paula MöllingList of authors in order
- Landing page
-
https://doi.org/10.1099/mgen.0.001124Publisher landing page
- Open access
-
YesWhether a free full text is available
- OA status
-
goldOpen access status per OpenAlex
- OA URL
-
https://doi.org/10.1099/mgen.0.001124Direct OA link when available
- Concepts
-
Neisseria meningitidis, Carriage, Biology, Single-nucleotide polymorphism, Genome-wide association study, SNP, Gene, Genetics, Neisseria, Meningococcal disease, Microbiology, Genotype, Medicine, Bacteria, PathologyTop concepts (fields/topics) attached by OpenAlex
- Cited by
-
5Total citation count in OpenAlex
- Citations by year (recent)
-
2025: 3, 2024: 2Per-year citation counts (last 5 years)
- References (count)
-
75Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4387893260 |
|---|---|
| doi | https://doi.org/10.1099/mgen.0.001124 |
| ids.doi | https://doi.org/10.1099/mgen.0.001124 |
| ids.pmid | https://pubmed.ncbi.nlm.nih.gov/37874326 |
| ids.openalex | https://openalex.org/W4387893260 |
| fwci | 1.35071936 |
| mesh[0].qualifier_ui | |
| mesh[0].descriptor_ui | D006801 |
| mesh[0].is_major_topic | False |
| mesh[0].qualifier_name | |
| mesh[0].descriptor_name | Humans |
| mesh[1].qualifier_ui | Q000235 |
| mesh[1].descriptor_ui | D009345 |
| mesh[1].is_major_topic | True |
| mesh[1].qualifier_name | genetics |
| mesh[1].descriptor_name | Neisseria meningitidis |
| mesh[2].qualifier_ui | |
| mesh[2].descriptor_ui | D055106 |
| mesh[2].is_major_topic | False |
| mesh[2].qualifier_name | |
| mesh[2].descriptor_name | Genome-Wide Association Study |
| mesh[3].qualifier_ui | |
| mesh[3].descriptor_ui | D008585 |
| mesh[3].is_major_topic | True |
| mesh[3].qualifier_name | |
| mesh[3].descriptor_name | Meningitis, Meningococcal |
| mesh[4].qualifier_ui | |
| mesh[4].descriptor_ui | D001435 |
| mesh[4].is_major_topic | True |
| mesh[4].qualifier_name | |
| mesh[4].descriptor_name | Bacteriophages |
| mesh[5].qualifier_ui | |
| mesh[5].descriptor_ui | D022021 |
| mesh[5].is_major_topic | False |
| mesh[5].qualifier_name | |
| mesh[5].descriptor_name | Tacrolimus Binding Proteins |
| mesh[6].qualifier_ui | |
| mesh[6].descriptor_ui | D006801 |
| mesh[6].is_major_topic | False |
| mesh[6].qualifier_name | |
| mesh[6].descriptor_name | Humans |
| mesh[7].qualifier_ui | Q000235 |
| mesh[7].descriptor_ui | D009345 |
| mesh[7].is_major_topic | True |
| mesh[7].qualifier_name | genetics |
| mesh[7].descriptor_name | Neisseria meningitidis |
| mesh[8].qualifier_ui | |
| mesh[8].descriptor_ui | D055106 |
| mesh[8].is_major_topic | False |
| mesh[8].qualifier_name | |
| mesh[8].descriptor_name | Genome-Wide Association Study |
| mesh[9].qualifier_ui | |
| mesh[9].descriptor_ui | D008585 |
| mesh[9].is_major_topic | True |
| mesh[9].qualifier_name | |
| mesh[9].descriptor_name | Meningitis, Meningococcal |
| mesh[10].qualifier_ui | |
| mesh[10].descriptor_ui | D001435 |
| mesh[10].is_major_topic | True |
| mesh[10].qualifier_name | |
| mesh[10].descriptor_name | Bacteriophages |
| mesh[11].qualifier_ui | |
| mesh[11].descriptor_ui | D022021 |
| mesh[11].is_major_topic | False |
| mesh[11].qualifier_name | |
| mesh[11].descriptor_name | Tacrolimus Binding Proteins |
| mesh[12].qualifier_ui | |
| mesh[12].descriptor_ui | D006801 |
| mesh[12].is_major_topic | False |
| mesh[12].qualifier_name | |
| mesh[12].descriptor_name | Humans |
| mesh[13].qualifier_ui | Q000235 |
| mesh[13].descriptor_ui | D009345 |
| mesh[13].is_major_topic | True |
| mesh[13].qualifier_name | genetics |
| mesh[13].descriptor_name | Neisseria meningitidis |
| mesh[14].qualifier_ui | |
| mesh[14].descriptor_ui | D055106 |
| mesh[14].is_major_topic | False |
| mesh[14].qualifier_name | |
| mesh[14].descriptor_name | Genome-Wide Association Study |
| mesh[15].qualifier_ui | |
| mesh[15].descriptor_ui | D008585 |
| mesh[15].is_major_topic | True |
| mesh[15].qualifier_name | |
| mesh[15].descriptor_name | Meningitis, Meningococcal |
| mesh[16].qualifier_ui | |
| mesh[16].descriptor_ui | D001435 |
| mesh[16].is_major_topic | True |
| mesh[16].qualifier_name | |
| mesh[16].descriptor_name | Bacteriophages |
| mesh[17].qualifier_ui | |
| mesh[17].descriptor_ui | D022021 |
| mesh[17].is_major_topic | False |
| mesh[17].qualifier_name | |
| mesh[17].descriptor_name | Tacrolimus Binding Proteins |
| type | article |
| title | Genetic variants linked to the phenotypic outcome of invasive disease and carriage of Neisseria meningitidis |
| awards[0].id | https://openalex.org/G7976122023 |
| awards[0].funder_id | https://openalex.org/F4320325654 |
| awards[0].display_name | |
| awards[0].funder_award_id | OLL-967424 |
| awards[0].funder_display_name | Region Örebro län |
| biblio.issue | 10 |
| biblio.volume | 9 |
| biblio.last_page | |
| biblio.first_page | |
| grants[0].funder | https://openalex.org/F4320325654 |
| grants[0].award_id | OLL-967424 |
| grants[0].funder_display_name | Region Örebro län |
| topics[0].id | https://openalex.org/T10873 |
| topics[0].field.id | https://openalex.org/fields/24 |
| topics[0].field.display_name | Immunology and Microbiology |
| topics[0].score | 0.9998999834060669 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/2404 |
| topics[0].subfield.display_name | Microbiology |
| topics[0].display_name | Bacterial Infections and Vaccines |
| topics[1].id | https://openalex.org/T10654 |
| topics[1].field.id | https://openalex.org/fields/27 |
| topics[1].field.display_name | Medicine |
| topics[1].score | 0.9970999956130981 |
| topics[1].domain.id | https://openalex.org/domains/4 |
| topics[1].domain.display_name | Health Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/2713 |
| topics[1].subfield.display_name | Epidemiology |
| topics[1].display_name | Pneumonia and Respiratory Infections |
| topics[2].id | https://openalex.org/T10015 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.9957000017166138 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Genomics and Phylogenetic Studies |
| funders[0].id | https://openalex.org/F4320325654 |
| funders[0].ror | |
| funders[0].display_name | Region Örebro län |
| is_xpac | False |
| apc_list.value | 1500 |
| apc_list.currency | GBP |
| apc_list.value_usd | 1839 |
| apc_paid.value | 2444 |
| apc_paid.currency | EUR |
| apc_paid.value_usd | 2635 |
| concepts[0].id | https://openalex.org/C2778458888 |
| concepts[0].level | 3 |
| concepts[0].score | 0.7261930108070374 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q154625 |
| concepts[0].display_name | Neisseria meningitidis |
| concepts[1].id | https://openalex.org/C32762888 |
| concepts[1].level | 2 |
| concepts[1].score | 0.7082807421684265 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q235356 |
| concepts[1].display_name | Carriage |
| concepts[2].id | https://openalex.org/C86803240 |
| concepts[2].level | 0 |
| concepts[2].score | 0.6696023941040039 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[2].display_name | Biology |
| concepts[3].id | https://openalex.org/C153209595 |
| concepts[3].level | 4 |
| concepts[3].score | 0.5744802951812744 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q501128 |
| concepts[3].display_name | Single-nucleotide polymorphism |
| concepts[4].id | https://openalex.org/C106208931 |
| concepts[4].level | 5 |
| concepts[4].score | 0.5278598666191101 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q1098876 |
| concepts[4].display_name | Genome-wide association study |
| concepts[5].id | https://openalex.org/C139275648 |
| concepts[5].level | 5 |
| concepts[5].score | 0.5146861672401428 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q17134011 |
| concepts[5].display_name | SNP |
| concepts[6].id | https://openalex.org/C104317684 |
| concepts[6].level | 2 |
| concepts[6].score | 0.505645751953125 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[6].display_name | Gene |
| concepts[7].id | https://openalex.org/C54355233 |
| concepts[7].level | 1 |
| concepts[7].score | 0.4845210909843445 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[7].display_name | Genetics |
| concepts[8].id | https://openalex.org/C2777954195 |
| concepts[8].level | 3 |
| concepts[8].score | 0.47862496972084045 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q132209 |
| concepts[8].display_name | Neisseria |
| concepts[9].id | https://openalex.org/C2776645655 |
| concepts[9].level | 4 |
| concepts[9].score | 0.4405049979686737 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q890785 |
| concepts[9].display_name | Meningococcal disease |
| concepts[10].id | https://openalex.org/C89423630 |
| concepts[10].level | 1 |
| concepts[10].score | 0.38777410984039307 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q7193 |
| concepts[10].display_name | Microbiology |
| concepts[11].id | https://openalex.org/C135763542 |
| concepts[11].level | 3 |
| concepts[11].score | 0.18620255589485168 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q106016 |
| concepts[11].display_name | Genotype |
| concepts[12].id | https://openalex.org/C71924100 |
| concepts[12].level | 0 |
| concepts[12].score | 0.1390003263950348 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q11190 |
| concepts[12].display_name | Medicine |
| concepts[13].id | https://openalex.org/C523546767 |
| concepts[13].level | 2 |
| concepts[13].score | 0.09085667133331299 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q10876 |
| concepts[13].display_name | Bacteria |
| concepts[14].id | https://openalex.org/C142724271 |
| concepts[14].level | 1 |
| concepts[14].score | 0.0 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q7208 |
| concepts[14].display_name | Pathology |
| keywords[0].id | https://openalex.org/keywords/neisseria-meningitidis |
| keywords[0].score | 0.7261930108070374 |
| keywords[0].display_name | Neisseria meningitidis |
| keywords[1].id | https://openalex.org/keywords/carriage |
| keywords[1].score | 0.7082807421684265 |
| keywords[1].display_name | Carriage |
| keywords[2].id | https://openalex.org/keywords/biology |
| keywords[2].score | 0.6696023941040039 |
| keywords[2].display_name | Biology |
| keywords[3].id | https://openalex.org/keywords/single-nucleotide-polymorphism |
| keywords[3].score | 0.5744802951812744 |
| keywords[3].display_name | Single-nucleotide polymorphism |
| keywords[4].id | https://openalex.org/keywords/genome-wide-association-study |
| keywords[4].score | 0.5278598666191101 |
| keywords[4].display_name | Genome-wide association study |
| keywords[5].id | https://openalex.org/keywords/snp |
| keywords[5].score | 0.5146861672401428 |
| keywords[5].display_name | SNP |
| keywords[6].id | https://openalex.org/keywords/gene |
| keywords[6].score | 0.505645751953125 |
| keywords[6].display_name | Gene |
| keywords[7].id | https://openalex.org/keywords/genetics |
| keywords[7].score | 0.4845210909843445 |
| keywords[7].display_name | Genetics |
| keywords[8].id | https://openalex.org/keywords/neisseria |
| keywords[8].score | 0.47862496972084045 |
| keywords[8].display_name | Neisseria |
| keywords[9].id | https://openalex.org/keywords/meningococcal-disease |
| keywords[9].score | 0.4405049979686737 |
| keywords[9].display_name | Meningococcal disease |
| keywords[10].id | https://openalex.org/keywords/microbiology |
| keywords[10].score | 0.38777410984039307 |
| keywords[10].display_name | Microbiology |
| keywords[11].id | https://openalex.org/keywords/genotype |
| keywords[11].score | 0.18620255589485168 |
| keywords[11].display_name | Genotype |
| keywords[12].id | https://openalex.org/keywords/medicine |
| keywords[12].score | 0.1390003263950348 |
| keywords[12].display_name | Medicine |
| keywords[13].id | https://openalex.org/keywords/bacteria |
| keywords[13].score | 0.09085667133331299 |
| keywords[13].display_name | Bacteria |
| language | en |
| locations[0].id | doi:10.1099/mgen.0.001124 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4210186362 |
| locations[0].source.issn | 2057-5858 |
| locations[0].source.type | journal |
| locations[0].source.is_oa | True |
| locations[0].source.issn_l | 2057-5858 |
| locations[0].source.is_core | True |
| locations[0].source.is_in_doaj | True |
| locations[0].source.display_name | Microbial Genomics |
| locations[0].source.host_organization | https://openalex.org/P4310320497 |
| locations[0].source.host_organization_name | Microbiology Society |
| locations[0].source.host_organization_lineage | https://openalex.org/P4310320497 |
| locations[0].source.host_organization_lineage_names | Microbiology Society |
| locations[0].license | cc-by |
| locations[0].pdf_url | |
| locations[0].version | publishedVersion |
| locations[0].raw_type | journal-article |
| locations[0].license_id | https://openalex.org/licenses/cc-by |
| locations[0].is_accepted | True |
| locations[0].is_published | True |
| locations[0].raw_source_name | Microbial Genomics |
| locations[0].landing_page_url | https://doi.org/10.1099/mgen.0.001124 |
| locations[1].id | pmid:37874326 |
| locations[1].is_oa | False |
| locations[1].source.id | https://openalex.org/S4306525036 |
| locations[1].source.issn | |
| locations[1].source.type | repository |
| locations[1].source.is_oa | False |
| locations[1].source.issn_l | |
| locations[1].source.is_core | False |
| locations[1].source.is_in_doaj | False |
| locations[1].source.display_name | PubMed |
| locations[1].source.host_organization | https://openalex.org/I1299303238 |
| locations[1].source.host_organization_name | National Institutes of Health |
| locations[1].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[1].license | |
| locations[1].pdf_url | |
| locations[1].version | publishedVersion |
| locations[1].raw_type | |
| locations[1].license_id | |
| locations[1].is_accepted | True |
| locations[1].is_published | True |
| locations[1].raw_source_name | Microbial genomics |
| locations[1].landing_page_url | https://pubmed.ncbi.nlm.nih.gov/37874326 |
| locations[2].id | pmh:oai:pubmedcentral.nih.gov:10634450 |
| locations[2].is_oa | True |
| locations[2].source.id | https://openalex.org/S2764455111 |
| locations[2].source.issn | |
| locations[2].source.type | repository |
| locations[2].source.is_oa | False |
| locations[2].source.issn_l | |
| locations[2].source.is_core | False |
| locations[2].source.is_in_doaj | False |
| locations[2].source.display_name | PubMed Central |
| locations[2].source.host_organization | https://openalex.org/I1299303238 |
| locations[2].source.host_organization_name | National Institutes of Health |
| locations[2].source.host_organization_lineage | https://openalex.org/I1299303238 |
| locations[2].license | cc-by |
| locations[2].pdf_url | https://pmc.ncbi.nlm.nih.gov/articles/PMC10634450/pdf/mgen-9-1124.pdf |
| locations[2].version | submittedVersion |
| locations[2].raw_type | Text |
| locations[2].license_id | https://openalex.org/licenses/cc-by |
| locations[2].is_accepted | False |
| locations[2].is_published | False |
| locations[2].raw_source_name | Microb Genom |
| locations[2].landing_page_url | https://www.ncbi.nlm.nih.gov/pmc/articles/10634450 |
| indexed_in | crossref, doaj, pubmed |
| authorships[0].author.id | https://openalex.org/A5017125617 |
| authorships[0].author.orcid | https://orcid.org/0009-0004-4040-4891 |
| authorships[0].author.display_name | Lorraine Eriksson |
| authorships[0].countries | SE |
| authorships[0].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[0].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[0].institutions[0].id | https://openalex.org/I26437253 |
| authorships[0].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[0].institutions[0].type | education |
| authorships[0].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[0].institutions[0].country_code | SE |
| authorships[0].institutions[0].display_name | Örebro University |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Lorraine Eriksson |
| authorships[0].is_corresponding | False |
| authorships[0].raw_affiliation_strings | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[1].author.id | https://openalex.org/A5014323760 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-4502-7883 |
| authorships[1].author.display_name | Thor Bech Johannesen |
| authorships[1].countries | DK |
| authorships[1].affiliations[0].institution_ids | https://openalex.org/I101527212 |
| authorships[1].affiliations[0].raw_affiliation_string | Department of Bacteria, Parasites and Fungi, Statens Serum Institut, Copenhagen, Denmark |
| authorships[1].institutions[0].id | https://openalex.org/I101527212 |
| authorships[1].institutions[0].ror | https://ror.org/0417ye583 |
| authorships[1].institutions[0].type | facility |
| authorships[1].institutions[0].lineage | https://openalex.org/I101527212 |
| authorships[1].institutions[0].country_code | DK |
| authorships[1].institutions[0].display_name | Statens Serum Institut |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Thor Bech Johannesen |
| authorships[1].is_corresponding | False |
| authorships[1].raw_affiliation_strings | Department of Bacteria, Parasites and Fungi, Statens Serum Institut, Copenhagen, Denmark |
| authorships[2].author.id | https://openalex.org/A5000577414 |
| authorships[2].author.orcid | https://orcid.org/0000-0003-4637-8626 |
| authorships[2].author.display_name | Bianca Stenmark |
| authorships[2].countries | SE |
| authorships[2].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[2].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[2].institutions[0].id | https://openalex.org/I26437253 |
| authorships[2].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[2].institutions[0].type | education |
| authorships[2].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[2].institutions[0].country_code | SE |
| authorships[2].institutions[0].display_name | Örebro University |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Bianca Stenmark |
| authorships[2].is_corresponding | False |
| authorships[2].raw_affiliation_strings | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[3].author.id | https://openalex.org/A5108454211 |
| authorships[3].author.orcid | |
| authorships[3].author.display_name | Susanne Jacobsson |
| authorships[3].countries | SE |
| authorships[3].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[3].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[3].institutions[0].id | https://openalex.org/I26437253 |
| authorships[3].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[3].institutions[0].type | education |
| authorships[3].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[3].institutions[0].country_code | SE |
| authorships[3].institutions[0].display_name | Örebro University |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Susanne Jacobsson |
| authorships[3].is_corresponding | False |
| authorships[3].raw_affiliation_strings | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[4].author.id | https://openalex.org/A5024142251 |
| authorships[4].author.orcid | https://orcid.org/0000-0003-1999-3383 |
| authorships[4].author.display_name | Olof Säll |
| authorships[4].countries | SE |
| authorships[4].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[4].affiliations[0].raw_affiliation_string | Department of Infectious Diseases, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[4].institutions[0].id | https://openalex.org/I26437253 |
| authorships[4].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[4].institutions[0].type | education |
| authorships[4].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[4].institutions[0].country_code | SE |
| authorships[4].institutions[0].display_name | Örebro University |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Olof Säll |
| authorships[4].is_corresponding | False |
| authorships[4].raw_affiliation_strings | Department of Infectious Diseases, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[5].author.id | https://openalex.org/A5041896336 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Sara Thulin Hedberg |
| authorships[5].countries | SE |
| authorships[5].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[5].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[5].institutions[0].id | https://openalex.org/I26437253 |
| authorships[5].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[5].institutions[0].type | education |
| authorships[5].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[5].institutions[0].country_code | SE |
| authorships[5].institutions[0].display_name | Örebro University |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Sara Thulin Hedberg |
| authorships[5].is_corresponding | False |
| authorships[5].raw_affiliation_strings | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[6].author.id | https://openalex.org/A5112895827 |
| authorships[6].author.orcid | |
| authorships[6].author.display_name | Hans Fredlund |
| authorships[6].countries | SE |
| authorships[6].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[6].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[6].institutions[0].id | https://openalex.org/I26437253 |
| authorships[6].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[6].institutions[0].type | education |
| authorships[6].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[6].institutions[0].country_code | SE |
| authorships[6].institutions[0].display_name | Örebro University |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Hans Fredlund |
| authorships[6].is_corresponding | False |
| authorships[6].raw_affiliation_strings | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[7].author.id | https://openalex.org/A5025613930 |
| authorships[7].author.orcid | https://orcid.org/0000-0003-0321-1180 |
| authorships[7].author.display_name | Marc Stegger |
| authorships[7].countries | DK, SE |
| authorships[7].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[7].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[7].affiliations[1].institution_ids | https://openalex.org/I101527212 |
| authorships[7].affiliations[1].raw_affiliation_string | Department of Bacteria, Parasites and Fungi, Statens Serum Institut, Copenhagen, Denmark |
| authorships[7].institutions[0].id | https://openalex.org/I101527212 |
| authorships[7].institutions[0].ror | https://ror.org/0417ye583 |
| authorships[7].institutions[0].type | facility |
| authorships[7].institutions[0].lineage | https://openalex.org/I101527212 |
| authorships[7].institutions[0].country_code | DK |
| authorships[7].institutions[0].display_name | Statens Serum Institut |
| authorships[7].institutions[1].id | https://openalex.org/I26437253 |
| authorships[7].institutions[1].ror | https://ror.org/05kytsw45 |
| authorships[7].institutions[1].type | education |
| authorships[7].institutions[1].lineage | https://openalex.org/I26437253 |
| authorships[7].institutions[1].country_code | SE |
| authorships[7].institutions[1].display_name | Örebro University |
| authorships[7].author_position | middle |
| authorships[7].raw_author_name | Marc Stegger |
| authorships[7].is_corresponding | False |
| authorships[7].raw_affiliation_strings | Department of Bacteria, Parasites and Fungi, Statens Serum Institut, Copenhagen, Denmark, Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[8].author.id | https://openalex.org/A5078151740 |
| authorships[8].author.orcid | https://orcid.org/0009-0000-1143-9105 |
| authorships[8].author.display_name | Paula Mölling |
| authorships[8].countries | SE |
| authorships[8].affiliations[0].institution_ids | https://openalex.org/I26437253 |
| authorships[8].affiliations[0].raw_affiliation_string | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| authorships[8].institutions[0].id | https://openalex.org/I26437253 |
| authorships[8].institutions[0].ror | https://ror.org/05kytsw45 |
| authorships[8].institutions[0].type | education |
| authorships[8].institutions[0].lineage | https://openalex.org/I26437253 |
| authorships[8].institutions[0].country_code | SE |
| authorships[8].institutions[0].display_name | Örebro University |
| authorships[8].author_position | last |
| authorships[8].raw_author_name | Paula Mölling |
| authorships[8].is_corresponding | False |
| authorships[8].raw_affiliation_strings | Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden |
| has_content.pdf | False |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://doi.org/10.1099/mgen.0.001124 |
| open_access.oa_status | gold |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-10-10T00:00:00 |
| display_name | Genetic variants linked to the phenotypic outcome of invasive disease and carriage of Neisseria meningitidis |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10873 |
| primary_topic.field.id | https://openalex.org/fields/24 |
| primary_topic.field.display_name | Immunology and Microbiology |
| primary_topic.score | 0.9998999834060669 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/2404 |
| primary_topic.subfield.display_name | Microbiology |
| primary_topic.display_name | Bacterial Infections and Vaccines |
| related_works | https://openalex.org/W1957800381, https://openalex.org/W2003004797, https://openalex.org/W2901294968, https://openalex.org/W2996668822, https://openalex.org/W2026960182, https://openalex.org/W2055298296, https://openalex.org/W2014354026, https://openalex.org/W2108591411, https://openalex.org/W2616329816, https://openalex.org/W2100149488 |
| cited_by_count | 5 |
| counts_by_year[0].year | 2025 |
| counts_by_year[0].cited_by_count | 3 |
| counts_by_year[1].year | 2024 |
| counts_by_year[1].cited_by_count | 2 |
| locations_count | 3 |
| best_oa_location.id | doi:10.1099/mgen.0.001124 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4210186362 |
| best_oa_location.source.issn | 2057-5858 |
| best_oa_location.source.type | journal |
| best_oa_location.source.is_oa | True |
| best_oa_location.source.issn_l | 2057-5858 |
| best_oa_location.source.is_core | True |
| best_oa_location.source.is_in_doaj | True |
| best_oa_location.source.display_name | Microbial Genomics |
| best_oa_location.source.host_organization | https://openalex.org/P4310320497 |
| best_oa_location.source.host_organization_name | Microbiology Society |
| best_oa_location.source.host_organization_lineage | https://openalex.org/P4310320497 |
| best_oa_location.source.host_organization_lineage_names | Microbiology Society |
| best_oa_location.license | cc-by |
| best_oa_location.pdf_url | |
| best_oa_location.version | publishedVersion |
| best_oa_location.raw_type | journal-article |
| best_oa_location.license_id | https://openalex.org/licenses/cc-by |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | True |
| best_oa_location.raw_source_name | Microbial Genomics |
| best_oa_location.landing_page_url | https://doi.org/10.1099/mgen.0.001124 |
| primary_location.id | doi:10.1099/mgen.0.001124 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4210186362 |
| primary_location.source.issn | 2057-5858 |
| primary_location.source.type | journal |
| primary_location.source.is_oa | True |
| primary_location.source.issn_l | 2057-5858 |
| primary_location.source.is_core | True |
| primary_location.source.is_in_doaj | True |
| primary_location.source.display_name | Microbial Genomics |
| primary_location.source.host_organization | https://openalex.org/P4310320497 |
| primary_location.source.host_organization_name | Microbiology Society |
| primary_location.source.host_organization_lineage | https://openalex.org/P4310320497 |
| primary_location.source.host_organization_lineage_names | Microbiology Society |
| primary_location.license | cc-by |
| primary_location.pdf_url | |
| primary_location.version | publishedVersion |
| primary_location.raw_type | journal-article |
| primary_location.license_id | https://openalex.org/licenses/cc-by |
| primary_location.is_accepted | True |
| primary_location.is_published | True |
| primary_location.raw_source_name | Microbial Genomics |
| primary_location.landing_page_url | https://doi.org/10.1099/mgen.0.001124 |
| publication_date | 2023-10-24 |
| publication_year | 2023 |
| referenced_works | https://openalex.org/W2133410828, https://openalex.org/W2987841563, https://openalex.org/W2013053657, https://openalex.org/W1991530099, https://openalex.org/W2031547878, https://openalex.org/W2414583777, https://openalex.org/W2916950596, https://openalex.org/W2089775764, https://openalex.org/W2142503351, https://openalex.org/W2969722206, https://openalex.org/W2162434670, https://openalex.org/W2986066723, https://openalex.org/W2605780312, https://openalex.org/W2137586712, https://openalex.org/W4320499374, https://openalex.org/W2956070303, https://openalex.org/W3007983701, https://openalex.org/W2167788433, https://openalex.org/W2949660875, https://openalex.org/W2556693791, https://openalex.org/W2952473089, https://openalex.org/W2784283722, https://openalex.org/W2159954944, https://openalex.org/W2472226552, https://openalex.org/W2141052558, https://openalex.org/W1972145956, https://openalex.org/W2927961455, https://openalex.org/W2120902911, https://openalex.org/W2096093282, https://openalex.org/W2122673596, https://openalex.org/W1886724263, https://openalex.org/W2043884503, https://openalex.org/W2891078715, https://openalex.org/W3004681462, https://openalex.org/W2045162565, https://openalex.org/W1577095452, https://openalex.org/W2097846683, https://openalex.org/W2772790023, https://openalex.org/W1990425530, https://openalex.org/W2158380232, https://openalex.org/W2126495901, https://openalex.org/W1906836500, https://openalex.org/W2108177341, https://openalex.org/W2156731936, https://openalex.org/W2174942393, https://openalex.org/W2110040181, https://openalex.org/W2735053618, https://openalex.org/W2473486641, https://openalex.org/W2069276860, https://openalex.org/W2040626353, https://openalex.org/W2130497375, https://openalex.org/W2104012811, https://openalex.org/W2162423297, https://openalex.org/W2528706897, https://openalex.org/W2026157295, https://openalex.org/W2143085076, https://openalex.org/W2012867961, https://openalex.org/W2134964275, https://openalex.org/W2082919450, https://openalex.org/W2132214292, https://openalex.org/W2001334035, https://openalex.org/W2069447355, https://openalex.org/W2115416724, https://openalex.org/W2112292543, https://openalex.org/W3207295113, https://openalex.org/W2087387372, https://openalex.org/W2144014919, https://openalex.org/W2736238444, https://openalex.org/W1970332030, https://openalex.org/W2890293221, https://openalex.org/W2159972665, https://openalex.org/W1995520553, https://openalex.org/W4307935500, https://openalex.org/W2047890226, https://openalex.org/W2748680502 |
| referenced_works_count | 75 |
| abstract_inverted_index.( | 79, 90 |
| abstract_inverted_index., | 100, 201, 226, 228, 232 |
| abstract_inverted_index./ | 178 |
| abstract_inverted_index.a | 4, 63, 140, 166, 194, 254 |
| abstract_inverted_index.n | 80, 91 |
| abstract_inverted_index.3, | 174 |
| abstract_inverted_index.In | 69, 222 |
| abstract_inverted_index.N. | 59, 73, 273, 291 |
| abstract_inverted_index.No | 26 |
| abstract_inverted_index.Of | 159 |
| abstract_inverted_index.as | 21 |
| abstract_inverted_index.at | 86 |
| abstract_inverted_index.be | 3, 49, 208 |
| abstract_inverted_index.in | 7, 77, 256 |
| abstract_inverted_index.is | 13 |
| abstract_inverted_index.of | 16, 169, 287, 290 |
| abstract_inverted_index.or | 271 |
| abstract_inverted_index.to | 33, 43, 51, 103, 207, 284, 296 |
| abstract_inverted_index.One | 110 |
| abstract_inverted_index.SNP | 115, 127, 148, 186 |
| abstract_inverted_index.The | 39, 96, 133, 185, 258 |
| abstract_inverted_index.aim | 40 |
| abstract_inverted_index.and | 24, 82, 108, 112, 121, 124, 144, 180, 203, 216, 230, 242, 293 |
| abstract_inverted_index.are | 266 |
| abstract_inverted_index.but | 12 |
| abstract_inverted_index.can | 2 |
| abstract_inverted_index.one | 113, 125 |
| abstract_inverted_index.our | 285 |
| abstract_inverted_index.the | 8, 145, 152, 160, 175, 223, 276, 288 |
| abstract_inverted_index.was | 42, 101, 190 |
| abstract_inverted_index.1-P) | 157 |
| abstract_inverted_index.GWAS | 97 |
| abstract_inverted_index.PilC | 229 |
| abstract_inverted_index.PilS | 200 |
| abstract_inverted_index.PorB | 172 |
| abstract_inverted_index.also | 14 |
| abstract_inverted_index.been | 31 |
| abstract_inverted_index.both | 240 |
| abstract_inverted_index.data | 259 |
| abstract_inverted_index.find | 44 |
| abstract_inverted_index.fkbp | 191, 225 |
| abstract_inverted_index.from | 36, 199, 214 |
| abstract_inverted_index.gene | 111, 134, 167 |
| abstract_inverted_index.glmU | 149, 227 |
| abstract_inverted_index.have | 30, 182 |
| abstract_inverted_index.help | 295 |
| abstract_inverted_index.here | 41, 261 |
| abstract_inverted_index.mtrD | 215 |
| abstract_inverted_index.pilE | 177, 231 |
| abstract_inverted_index.pilS | 179 |
| abstract_inverted_index.play | 253 |
| abstract_inverted_index.porB | 170 |
| abstract_inverted_index.role | 255 |
| abstract_inverted_index.such | 20 |
| abstract_inverted_index.tbpA | 217 |
| abstract_inverted_index.tbpB | 202 |
| abstract_inverted_index.that | 47, 236, 246, 265 |
| abstract_inverted_index.this | 70 |
| abstract_inverted_index.tspB | 181, 204 |
| abstract_inverted_index.were | 94, 116, 128, 205, 218, 234, 237 |
| abstract_inverted_index.with | 55, 118, 130, 136, 163, 188, 210, 220, 239, 269 |
| abstract_inverted_index.=103) | 81 |
| abstract_inverted_index.=213) | 92 |
| abstract_inverted_index.D33N, | 192 |
| abstract_inverted_index.S373C | 150 |
| abstract_inverted_index.These | 279 |
| abstract_inverted_index.class | 173 |
| abstract_inverted_index.could | 48, 252, 281, 294 |
| abstract_inverted_index.found | 206, 235 |
| abstract_inverted_index.genes | 107, 123, 161, 176, 224, 251 |
| abstract_inverted_index.human | 5 |
| abstract_inverted_index.known | 183 |
| abstract_inverted_index.phage | 141 |
| abstract_inverted_index.prove | 282 |
| abstract_inverted_index.seven | 122 |
| abstract_inverted_index.study | 67 |
| abstract_inverted_index.these | 250 |
| abstract_inverted_index.tract | 11 |
| abstract_inverted_index.upper | 9 |
| abstract_inverted_index.while | 212 |
| abstract_inverted_index.K-mers | 198 |
| abstract_inverted_index.Sweden | 78, 89 |
| abstract_inverted_index.across | 275 |
| abstract_inverted_index.enzyme | 153 |
| abstract_inverted_index.future | 298 |
| abstract_inverted_index.k-mers | 213, 233 |
| abstract_inverted_index.linked | 50 |
| abstract_inverted_index.study, | 71 |
| abstract_inverted_index.traits | 46, 264, 280 |
| abstract_inverted_index.versus | 57 |
| abstract_inverted_index.within | 249 |
| abstract_inverted_index.(FKBP). | 197 |
| abstract_inverted_index.(GWAS). | 68 |
| abstract_inverted_index.(GlcNAC | 156 |
| abstract_inverted_index.(SNPs), | 106 |
| abstract_inverted_index.applied | 102 |
| abstract_inverted_index.capable | 15 |
| abstract_inverted_index.causing | 17 |
| abstract_inverted_index.disease | 37, 61, 120, 138 |
| abstract_inverted_index.encodes | 139, 151 |
| abstract_inverted_index.genetic | 28, 45, 263 |
| abstract_inverted_index.k-mers. | 109 |
| abstract_inverted_index.markers | 29 |
| abstract_inverted_index.protein | 196 |
| abstract_inverted_index.species | 277 |
| abstract_inverted_index.through | 62 |
| abstract_inverted_index.treeWAS | 99 |
| abstract_inverted_index.vaccine | 299 |
| abstract_inverted_index.variant | 168 |
| abstract_inverted_index.Örebro | 87 |
| abstract_inverted_index.carriage | 35, 56, 83, 131, 164, 189, 241, 272 |
| abstract_inverted_index.detected | 32 |
| abstract_inverted_index.diseases | 19 |
| abstract_inverted_index.encoding | 171, 193 |
| abstract_inverted_index.identify | 297 |
| abstract_inverted_index.invasive | 18, 58, 72, 119, 137, 147, 243, 270 |
| abstract_inverted_index.isolates | 75, 84 |
| abstract_inverted_index.outcomes | 53 |
| abstract_inverted_index.specific | 27, 247 |
| abstract_inverted_index.targets. | 300 |
| abstract_inverted_index.Neisseria | 0 |
| abstract_inverted_index.analysed. | 95 |
| abstract_inverted_index.analysis, | 98 |
| abstract_inverted_index.bacterial | 64 |
| abstract_inverted_index.carriage, | 211 |
| abstract_inverted_index.collected | 76, 85 |
| abstract_inverted_index.commensal | 6 |
| abstract_inverted_index.essential | 283 |
| abstract_inverted_index.highlight | 262 |
| abstract_inverted_index.isolates, | 165, 244 |
| abstract_inverted_index.isolates. | 38, 132 |
| abstract_inverted_index.presented | 260 |
| abstract_inverted_index.associated | 54, 117, 129, 135, 146, 162, 187, 209, 219, 238, 268 |
| abstract_inverted_index.functions. | 184 |
| abstract_inverted_index.indicating | 245 |
| abstract_inverted_index.meningitis | 23 |
| abstract_inverted_index.phenotypic | 52 |
| abstract_inverted_index.variations | 248 |
| abstract_inverted_index.(NEIS1048), | 143 |
| abstract_inverted_index.1-phosphate | 155 |
| abstract_inverted_index.2018–2019 | 93 |
| abstract_inverted_index.University, | 88 |
| abstract_inverted_index.association | 66 |
| abstract_inverted_index.distinguish | 34 |
| abstract_inverted_index.genome-wide | 65 |
| abstract_inverted_index.population. | 278 |
| abstract_inverted_index.respiratory | 10 |
| abstract_inverted_index.transposase | 142 |
| abstract_inverted_index.meningitidis | 1, 60, 74, 274, 292 |
| abstract_inverted_index.septicaemia. | 25 |
| abstract_inverted_index.FK506-binding | 195 |
| abstract_inverted_index.invasiveness. | 221, 257 |
| abstract_inverted_index.meningococcal | 22 |
| abstract_inverted_index.pathogenicity | 289 |
| abstract_inverted_index.polymorphisms | 105 |
| abstract_inverted_index.significantly | 267 |
| abstract_inverted_index.understanding | 286 |
| abstract_inverted_index.non-synonymous | 114, 126 |
| abstract_inverted_index.single-nucleotide | 104 |
| abstract_inverted_index.uridyltransferase. | 158 |
| abstract_inverted_index.N-acetylglucosamine | 154 |
| cited_by_percentile_year.max | 97 |
| cited_by_percentile_year.min | 94 |
| countries_distinct_count | 2 |
| institutions_distinct_count | 9 |
| citation_normalized_percentile.value | 0.76355391 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |